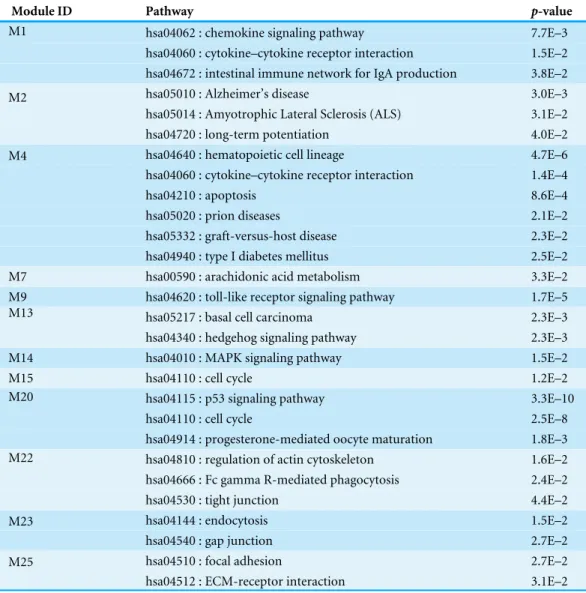
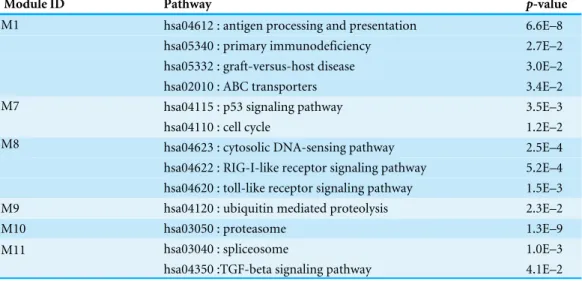
Network-based analysis of differentially expressed genes in cerebrospinal fluid (CSF) and blood reveals new candidate genes for multiple sclerosis
Texto
Imagem

Documentos relacionados
In the present study, we identified ubiquitous and tissue-specific [Na + ] i /[K + ] i -sensitive transcriptomes by comparative analysis of differentially expressed genes in
Comparing AA to CA astrocytes, differentially expressed genes that are associated with cell adhesion were ephrin B2 and GPR56, which were both upregulated, and ITGA6, which was
Whereas interactome guided analysis of differentially expressed genes uncovered several pathophysiologically relevant pathways in female visceral adipose and male skeletal muscle,
We identified 2418 mRNAs and 19 miRNAs as significantly differentially expressed, over 700 gene ontology (GO) terms and 83 KEGG pathways that were significantly enriched in GHS
sürdürmede etkili olmaktadır.[41] Evliler için ise alkol bağımlılığının gelişme riski daha yüksek bulunmakla birlikte, bağımlılığın sürme olasılığı bekarlara, dullara
Ingenuity pathway analysis was used to assemble a network based upon 13 differentially expressed focus genes and gene products that were upregulated in male VICs compared to
Furthermore, functional annotation, gene ontology enrichment, pathway enrichment and interaction network analyses were conducted on the differentially expressed genes (DEGs)
By com- paring gene expression levels between salt treatment and the control at each time point, 163 genes were identified to be differentially expressed in response to salt