Gene expression pro filing analysis of lung adenocarcinoma
Texto
Imagem

Documentos relacionados
Gene expression profiling showing differentially expressed genes between TTP -high and TTP -low expressing tumors in TCGA breast cancer ( A ), lung adenocarcinoma ( B ), lung
Our microarray analysis of the gene expression pro- file of degenerated IVD detected hundreds of differentially expressed genes that may be associated with IVD degener-
Key words: Differentially expressed proteins; Chronic atrophic gastritis; Proteomic study; Ribosomal protein S12 (RPS12); Proteasome activator subunit 1
Gene ontology (GO) enrichment analysis for the differentially expressed target genes of selected miRNAs in esophageal adenocarcinoma (EAC) and squamous-cell carcinoma (ESCC)..
We performed a larger scale interaction analysis between all pairs of SNPs that (i) are found in interacting genes according to a curated human protein-protein interaction network ( ,
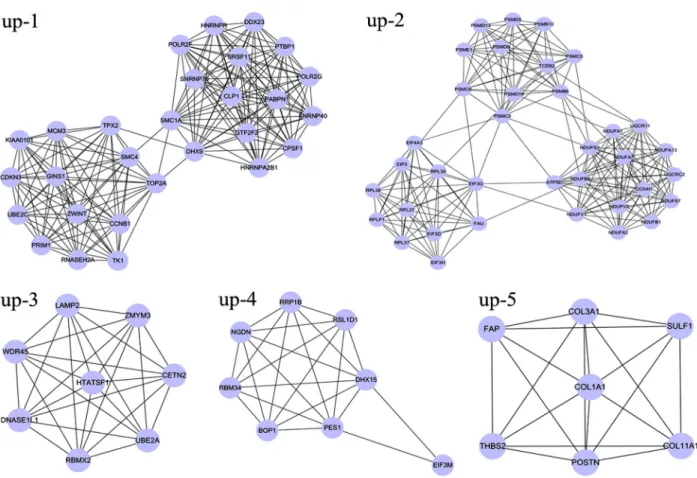
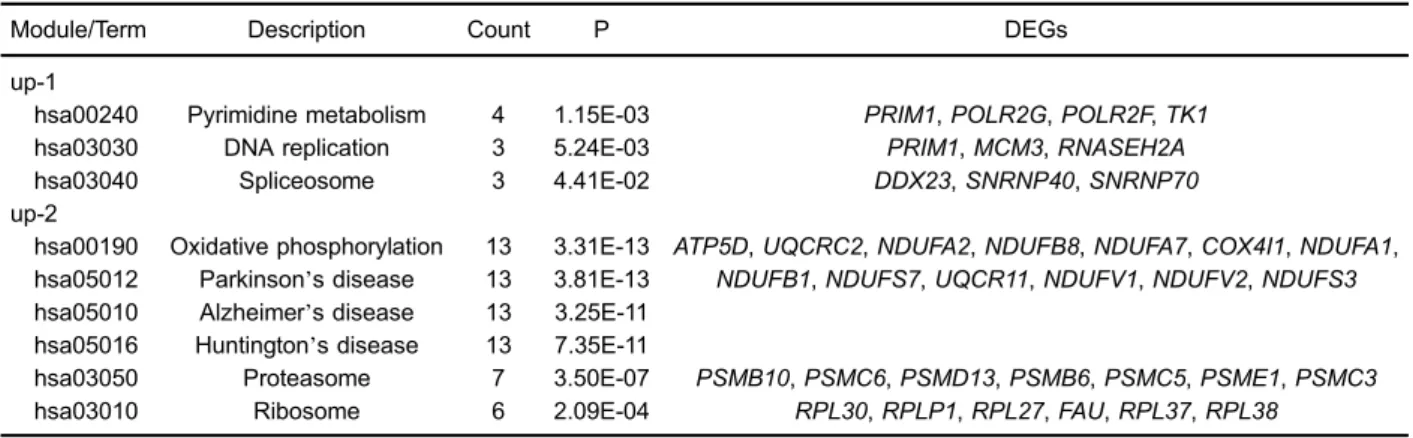
Furthermore, functional annotation, gene ontology enrichment, pathway enrichment and interaction network analyses were conducted on the differentially expressed genes (DEGs)
Gene ontology (GO) enrichment of differentially expressed genes revealed protein folding, cell morphogenesis and cellular component morphogenesis as the top three functional terms
By integrating gene expression signatures and somatic mutation data on a protein- protein interaction network, we show that the CRC recurrence phenotype involves the dysregulation