Molecular evolution of the ent -kaurenoic acid oxidase gene in Oryzeae
Texto
Imagem


Documentos relacionados
Flavonoid content correlated positively with the honey color, melanoidins and phenol content but negatively with the IC 50 values of the scavenging capacity of ABTS free rad-..
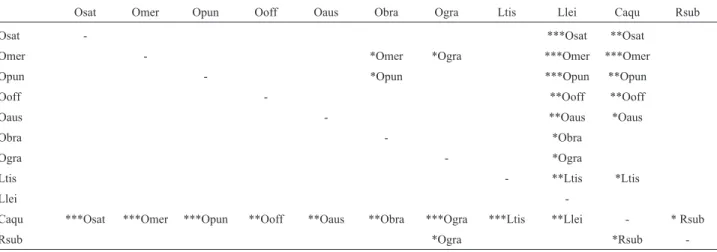
Index terms: Annonaceae, Branch lengths, Codon models, Non-synonymous substitution rate, Phylogenetics, Synonymous substitution rate.. 1 Palestra Anonáceas - V Congresso
The wheat yield was negatively correlated with soil penetration resistance and bulk density, and positively correlated with the gravimetric water content in the field, total
Segundo Stojanovic 2006, a implementação de programas de identificação de talentos pode trazer bastantes benefícios: 1 Em geral, os programas de identificação de talentos promovem
Three measures for the degree of codon usage bias indicated the presence of bias in all the sequences, suggesting high expression levels in all the genes.. Protein structures
The soluble PNAs content showed no significant correlation with protein, oil, sucrose and raffinose + stachyose contents, but oligosaccharides showed a negative correlation
With the calcium silicate source, the severity of brown spot and the panicle blast incidence were negatively correlated (p < 0.01) with the content of silicon in the plants,
descriptive statistics such as nucleotide, amino acid, and codon frequencies, dinu- cleotide and diamino acid frequencies, analysis of codon usage and amino acid usage bias; and