http://dx.doi.org/10.1590/1806-9061-2017-0662
Author(s)
Wang QI,II Yuan XI
Li CI Deng HI Wang CI
I College of animal science, Fujian agricultu-re and foagricultu-restry university, Fuzhou, 350002, China;
II Fujian key laboratory of traditional Chine-se veterinary medicine and animal health (Fujian Agriculture and Forestry University) Fuzhou, Fujian 350002, China.
Mail Address
Corresponding author e-mail address Changkang Wang
Fujian Agriculture and Foresty University, Fuzhou, Fuzhou, Fujian, 350002, China. Tel: 086-0591-83758852
Email: Wangchangkangcn@163.com
Keywords
Yellow-feathered broiler, Interferon, Interferon-induced transmembrane protein 3, Gene cloning, Prokaryotic expression, Avian Reovirus.
Submitted: 10/October/2017 Approved: 08/January/2018
ABSTRACT
This study was carried to express
theinterferon-induced
transmembrane protein 3 (
IFITM3
)
invitro and examine its
function in inhibition of avian reovirus (ARV) replication. The
recombinant prokaryotic vector expressing yellow-feathered
broiler
IFITM3
was successfully constructed, and the recombinant
protein was expressed in competent
Escherichia coli
BL21 cells.
New Zealand white rabbits were immunized with the purified
recombinant protein to prepare a polyclonal antibody, with a titer
of 1:128,000. Immunohistochemistry, reverse transcription–PCR,
and real-time
fluorescence quantitative PCR showed that
IFITM3
was distributed in the yellow-feathered broiler immune organs,
and the expression of
IFITM3
in bursa of Fabricius was more than
in spleen and thymus. It was found that in the thymus, spleen and
bursa of Fabricius the mRNA expression levels of IFN
α
and
IFITM3
were significantly induced after ARV infection. And it was also
certified in the chicken embryo fibroblasts (CEFs) which infected
with ARV. Then the
IFN
α
was added into the cell culture medium
before CEFs were infected with ARV. The results indicated that
the mRNA of
IFITM3
expression was significantly increased and
ARV multiplication was significantly inhibited. And when the
expression of
IFITM3
was knocked down by siRNA-
IFITM3
, the
expression of
IFITM3
was significantly reduced, but the ARV
multiplication was significantly increased, which indicated that
IFITM3
protein could inhibit the ARV replication.
Abbreviations: avian reovirus, ARV; complementary deoxyribonucleic acid (cDNA); chicken embryonic fibroblasts, CEFs; double distilled water (ddH2O); Dulbecco’s modified Eagle medium, DMEM; glyceraldehyde-3-phosphate dehydrogenase gene, GAPDH; interferon alpha, IFNα; interferon-induced transmembrane protein, IFITM; intravenous, iv; median tissue culture infective dose ,TCID50; multiplicity of infection, MOI; National Center of Biotechnology Information, NCBI; phosphate-buffered saline, PBS; polymerase chain reaction, PCR; ribonucleic acid, RNA; reverse transcription, RT; sodium dodecyl sulfate polyacrylamide gel electrophoresis, SDS-PAGE; small interfering RNAs (siRNAs).
INTRODUCTION
antiviral pathways by inducing the expression of interferon-stimulated genes (ISGs) (Randall et al., 2008). The interferon-inducible transmembrane (IFITM) proteins were first identified in IFN-treated neuroblastoma cells in 1984, and are encoded by a class of ISGs (Chen et al., 1984). The IFITM gene family includes IFITM1, IFITM2, IFITM3, IFITM5, IFITM6,
IFITM7, and IFITM10 genes and some IFITM-like genes. These genes are induced by IFN and are widely expressed in tissues and organs, except IFITM5, which is only expressed in bone cells (Siegrist et al., 2011). The biological functions of IFITM3 include the inhibition of both viral replication and cell entry. Brass et al. (2009) demonstrated that the IFITM proteins inhibit the early replication of flaviviruses, including Dengue virus and
West Nile virus. Lu et al. (2009) confirmed that IFITM proteins inhibit HIV-1 replication, at least partly by interfering with viral entry.
Infectious diseases greatly influence poultry production, especially viral infections that induce im-mune suppression. Avian reovirus (ARV) infections affect the poultry industry throughout the world (Lorena et al., 2009). ARV causes viral tenosynovitis, malabsorption, and respiratory diseases in chickens (Teng et al., 2013), and ARV-induced immunosuppression is one of the most important pathogenic mechanisms of the virus (Chen et al., 2015). However, how the innate immunity of chickens plays its roles during ARV infection requires further study. Therefore, in this study, the IFITM3 gene of the yellow-feathered broiler was cloned, expressed, and confirmed, and the distribution of its expression in the chicken immune organs was investigated. And IFITM3 could inhibit ARV replication in vivo and in vitro that also was carried out.
MATERIALS AND METHODS
Materials
Virus
Avian reovirus strain S1133 was propagated in chicken embryonic fibroblasts (CEFs), which were cultured in RPMI 1640 medium supplemented with 10% foetal bovine serum, as previously described (Wang et al., 2012). The median tissue culture infective dose (TCID50) was 10−5.40 (multiplicity of infection [MOI]
= 8/mL).
Cells
CEFs were grown in Dulbecco’s modified Eagle medium (DMEM) containing streptomycin (50 μg/ml), penicillin G50 (50 μg/ml), and 5% foetal bovine serum, at 37 °C in 5% CO2.
Vectors
The vectors used were the pGEM®-T Easy vector (Promega, Wisconsin, USA) and pET-32a(+) (Promega, Wisconsin, USA).
Main reagents
The main reagents used included the TIANamp® Viral Total Nucleic Acid Purification Kit (TIANamp, Beijing, China); the GoScript™ Reverse Transcription System (Promega, Wisconsin, USA)); HilyMax™ Transfection Reagent (Dojindo, Shanghai, China); horseradish-peroxidase-labelled sheep anti-chick IgG antibody;
Escherichia coli DH5α cells (Promega, Wisconsin, USA);
E. coli BL21(Promega, Wisconsin, USA).
Total RNA extraction and cDNA amplification
Tissue samples (spleen, thymus, and bursa of Fabricius) were collected from healthy 20-day-old yellow-feathered broilers. Total RNA was extracted from the yellow-feathered broiler spleen with the TRIzol Plus RNA Purification Kit (TIANamp, Beijing, China). The first-strand cDNA was synthesized according to the manufacturer’s instructions. The total volume of the reverse transcription reaction mixture was 20 μl. The reaction was performed at 45 °C for 15 min and 72 °C for 15 min.
Primers design
Oligonucleotide primers for the PCR reactions and real time-PCR were designed (Table1). BstZ1restriction sites were incorporated at the 5¢ ends of the IFITM3
gene primers.
Cloning the chicken IFITM3 gene
The PCR product amplified with the specific primers was purified and cloned into the pGEM®-T Easy vector (Promega, Wisconsin, USA). This recombinant plasmid was designated pGEM-T–IFITM3 and transformed into competent E. coli DH5α cells. Positive clones were selected with blue–white selection, and the positive plasmids were confirmed with PCR and sequencing.
The IFITM3 sequence was compared using the National Center for Biotechnology Information (NCBI) online BLAST tool. The structure of the IFITM3 protein was predicted with the DNAMAN software. The palmitoylation sites were determined with the CSS-Palm2.0 software.
Construction of expression plasmids and
expression of pEASY™-E1–IFITM3
expression vector. The recombinant plasmid was designated PEASY™-E1–IFITM3 and was transformed into competent E. coli DH5α cells. Positive clones were selected with blue–white selection and confirmed with PCR (using the T7 upstream primer and the target gene downstream primer), sequencing, and restriction enzyme digestion. And PEASY™-E1–IFITM3
was transformed into Competent E. coli BL21(DE3) pLysS cells.
The pEASY™-E1–IFITM3-positive BL21(DE3) pLysS cells were cultured for 14 h at 37 ºC with 1 mmoL/mL IPTG (Transgen Biotech, Beijing, China). The bacterial cells were collected and centrifuged at 12,000 g for 1 min. The pellet was then re-suspended in phosphate buffer (PBS) and broken with ultrasonography. The protein was denatured and identified with SDS-PAGE.
Purification of recombinant IFITM3
(rIFITM3) and preparation of polyclonal antibody
The recombinant protein was purified with affinity chromatography on ProteinIso™ Ni-NTA Resin (Transgen Biotech, Beijing, China). rIFITM3 was identified with SDS-PAGE and Western blotting.
An anti-rIFITM3 polyclonal antibody was prepared as described previously (Zheng et al., 2014). Three New Zealand white rabbits were inoculated by subcutaneous injection with 1 mL (1 mg/mL) of purified IFITM protein, which was emulsified with an equal amount of Freund’s complete adjuvant. Immunization was boosted twice by inoculation with 1 mL of antigen mixed with an equal volume Freund’s incomplete adjuvant at 2-weekly intervals. The fourth immunization was performed 7 days after the third immunization, with 1 mL (1 mg/mL) of purified IFITM protein by intravenous (iv) injection.
The antibody titer was determined with the enzyme-linked immunosorbent assay.
I m m u n o h i s t o c h e m i c a l localization of chicken IFITM3 in immune organs
The tissue samples including thymus, spleen and bursa of Fabricius, were dehydrated in a graded series of ethanol, embedded in paraffin, and sectioned at 4–6 μm with a histotome. After dewaxing and hydration, the sections were analysed with immunohistochemistry method. For immunohistochemical analysis, the sections were incubated with the rabbit anti-IFITM3 antibody at 37 °C for 60-120 min. After rinsed with PBS for 5 min three times, the sections were incubated with a biotin-conjugated goat anti-rabbit antibody (Keygen Biotech Co. Ltd, Nanjing, China) at 37 °C for 10-30 min. After the sections were washed, they were treated with streptavidin–peroxidase for 10 min and then with a diaminobenzidine solution. The sections were then counterstained with haematoxylin and mounted with neutral gum.
IFITM3 mRNA levels in chicken immune
organs
IFITM3 mRNA levels in chicken immune organs were examined by RT-PCR and SYBR Green real-time PCR. RT-PCR was performed with 1 mL of cDNA, 0.5
mL of each primer, 1 mL of PCR mix, and 8 ml of double distilled water (ddH2O), in a total volume of 20 mL. The cycling conditions for amplification were 95 °C for 5 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, followed by a final extension at 72 °C for 10 min. The products were analysed with electrophoresis on 1.0% agarose gel and sequenced by Dalian TaKaRa Company (Dalian,China).
The IFITM3 mRNA levels in the chicken immune organs were detected with SYBR Green real-time
fluorescence quantitative
PCR. Specific primers were designed (Table 1), and were synthesized by Invitrogen Company. Reaction volumes of 20 μL contained 10 μL of SYBR® Premix Ex Taq™ (2×) (Promega), 0.4 μLof FastStart Universal Probe Master, 0.4 μL of forward primer (10 μM), 0.4 μL of reverse primer (10 μM), 2 μL of cDNA, and 6.8 μL of RNase-free ddH2O. The PCR cycling conditions were 45 cycles of initial denaturation at 95 °C for 30 s, 95 °C for 5 s, and 60 °C for 34 s. The data were analysed with the ΔCT method using the following formula: target gene expression = 2−ΔCt
Table 1 – Primer sequences
Gene name Primer sequences (5’-3’) Amplification
fragments(bp) IFITM3-F
IFITM3-R
5’ATGCAGAGCTACCCTCAGCA3’
5’TCAGGGCCGCACAGTGTACAA3’ 342
IFITM3 (BstZ1)-F IFITM3 (BstZ1)-R
5’ATTCGGCCGATGCAGAGCTACCCTCAGCA3’ 5’TAAGCCGGCTCAGGGCCGCACAGTGTACAA3’ 360 IFITM3-F IFITM3-R 5’CAGAGCTACCCTCAGCACAC -3’ 5’CTGTCCTCCCATAGCTGCTG-3’ 235
IFNα-F IFNα-R
5’ATGGCTGTGCCTGCAAGC -3’
5’TTGCCTGTGAGGTTGTGGAT-3’ 569
IFNα-F IFNα-R
(ΔCt = Cttarget gene – Ctreference gene). The glyceraldehyde-3-phosphate dehydrogenase gene (GAPDH) was used as the reference gene. Statistical differences were determined with t test in SPSS 17.0.
IFITM3 gene expression in ARV-infected
chickens immune organs and CEF
Thirty 20-day-old yellow-feathered broilers were randomly divided into the control group (five and the experimental group) tweenty five. The experimental group chickens were infected with 0.2 mL ARV, andnd five control chickens were slaughtered at the first day of ARV iontreatment. Then five infection ducks were slaughtered every after every infection. The thymus, spleen, and bursa of Fabricius were collected from each broiler. The expression of IFTIM3 and IFNα and the replication of ARV were detected using reverse transcription (RT)–PCR and SYBR real-time fluorescence quantitative PCR.
CEFs grew in six-well tissue-culture plates with MDEM cell medium for 37 °C 24 h. CEF cells were collected after inoculation with ARV 0 h, 12 h, 24 h, 48 h, 72 h respectively. The cells were t digested with 0.25% pancreatic enzyme. The total RNA was extracted and the mRNA expression of IFN, IFTIM3 and ARV was determined with PCR and SYBR real-time fluorescence quantitative PCR.
Analysis of IFN sensitivity
CEFs grown in six-well tissue-culture plates were either mock-treated or treated with IFNα (100 IU/ml) for 0.5 h at 37 °C before viral inoculation. Then, these cells were challenged with 0.1mL of ARV for 24 h and digested with 0.25% pancreatic enzyme. The total RNA was extracted and the expression of IFTIM3 was determined with PCR.
IFITM3-directed small interfering RNAs (siRNAs) were designed (F: 5¢-GAGCCUACGAAACCUUAAUdTdT-3¢
and R: 5¢-AUUAAGGUUUCGUAGGCUCdTdT-3¢) and
transfected into CEF cells. The cells were inoculated with 0.1mL of ARV for 24 h and then digested with 0.25% pancreatic enzyme. The total RNA was extracted and the mRNA expressions of IFNα, IFTIM3 and ARV were detected with PCR and SYBR real-time
fluorescence quantitative
PCR.RESULTS
Cloning and analysis of the IFITM3 gene
Total RNA
was
extracted from the spleens of chickens and reversed transcribed to cDNA. PCR withspecific
primers amplified the 342-bp target gene (Figure 1A). The PCR
product
was ligated to the linearized pGEM-T Easycloning vector and competentE. coli DH5α cell were transfected with the recombinant plasmid. The recombinant plasmid was then identified with
enzymatic digestion. T
he target gene was 342 bp on 1% agarose gel electrophoresis (Figure 1B), indicating that the cloningvector was successfully constructed. The sequence of IFITM3 in the yellow-feathered broiler was determined and compared with homologous sequences using BLAST. The sequence of
IFITM3 in the yellow-feathered broiler was high ratio of homology to GallusIFITM3 (NCBI LOC422993).
Figura 1 – Parte A: PCR with specific primers amplified the 342-bp target gene. Parte B: The target gene was 342 bp on 1% agarose gel electrophoresis, indicating that the cloning vector was successfully constructed. Parte C: The amplified fragment was consis-tent with the anticipated fragment (584 bp).
The
IFITM3 gene sequences of Homo sapiens,Pan troglodytes,Macaca mulatta, Mus musculus,Sus scrofa,Bostaurus,Equuscaballus, Feliscatus, and the white-throated sparrow (Zonotrichia albicollis) were downloaded from NCBI
and their
homology was analysed with the DNAMAN software (Figure 2, Table 2). The evolutionary distance between the yellow-feathered broiler and Zonotrichia albicollis sequences was smallest, whereas the yellow-feathered broiler sequence was distant from the mammal sequences, andmost distant from
the human sequence.Table 2 – The evolutionary distance of IFITM3 gene in yellow-feathered broilers and other species
Species Evolutionary distance
Homo sapiens 0.059
Pan troglodytes 0.061
Macaca mulatta 0.104
Mus musculus 0.183
Sus scrofa 0.102
Bos taurus 0.114
Equus caballus 0.114
Felis catus 0.120
Amino acid sequence prediction and
analysis of the properties of IFITM3
Figura 2 – The IFITM3 gene sequences of Homo sapiens, Pan troglodytes, Macaca mulatta, Mus musculus, Sus scrofa, Bos taurus, Equus caballus, Felis catus, and the white-throated sparrow.
The amino acid sequence of IFITM3 was predicted and the protein properties were analyzed with DNAMAN. The isoelectric point (pI)
of
IFITM3
is 8.8, and its secondary structure consists of helices, folds,and curls, but predominantly folds and curls (Figure 3A). IFITM3 contains 113 amino acids, and has 20 fewer amino acids than human IFITM3. It contains a CD225 domain (Figure 2B), as does human IFITM3. There are two
transmembrane domains, at amino acids 37–65 and amino acids 78–106 (Figure 3C), and three antigen
ic
peptides. The IFITM3 protein of the yellow-feathered broileris hydrophobic (Figure 3 D).The S-palmitoylation
modification sites of IFITM3
in the yellow-feathered broiler were analyzed with the CSS
-Pal
M 2.0 software.
There are
four
potentialpalmitoylation sites at cysteines 45, 49, 53, and 89 (Table 3).
Table 3 – The IFITM3 protein of yellow-feathered broilers’S- palmitoylation modification site
Site Locus Possibility of palm acylating modified loci
49 NAFCLGL 0.65
89 FAFCVGL 0.61
53 LGLCALS 0.50
45 FVLCNAF 0.17
Construction of pEASY™-E1–cIFITM3 and
prokaryotic expression of the recombinant protein
The recombinant plasmid was amplified and identified with PCR using the T7 primer and target gene primer. The amplified fragment was consistent with the anticipated fragment (584 bp; Figure 1C.). The positive plasmid was sequenced by the Dalian TaKaKa Biotechnology Company, confirming the successful construction of the prokaryotic expression vector.
SDS-PAGE showed that the recombinant protein was successfully expressed in the BL21(DE3)pLysS cells, with the expected molecular weight of 11.5 kDa (Figure 4A). The recombinant protein was purified with affinity chromatography using ProteinIso™ Ni-NTA Resin. SDS-PAGE confirmed the successful purification of the recombinant protein (Figure 4B).
Specificity of the anti-IFITM3 polyclonal
antibody
After the immunization program, the anti-rIFITM3
rabbit serum was collected and the antibody titer was determined with ELISA method. The antibody titer reached to 1:256000. The rIFITM3 protein was
detected with western blotting, demonstrating that the polyclonal antibody was specific for the rIFITM3
protein (Fig. 4C).
Figura 4 – Parte A: SDS-PAGE showed that the recombinant protein was successfully expressed in the BL21(DE3)pLysS cells, with the expected molecular weight of 11.5 kDa. Parte B: The recombinant protein was purified with affinity chromatography using Protei-nIso™ Ni-NTA Resin. SDS-PAGE confirmed the successful purification of the recombi-nant protein. Parte C: The rIFITM3 protein was detected with western blotting, demons-trating that the polyclonal antibody was specific for the rIFITM3 protein.
Expression of IFITM3 in immune tissues
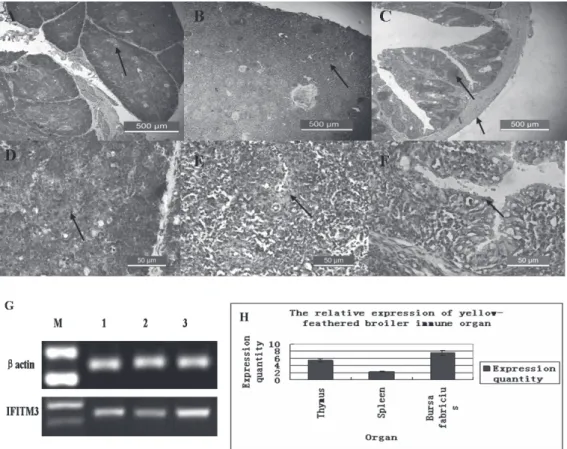
The distribution of IFITM3 in the chicken immune organs was examined with immunohistochemistry.
IFITM3 was distributed in the thymus (Figure 5A), spleen (Figure 5B), and bursa of Fabricius (Figure 5C). Positive cells were mainly distributed in the medulla of
the thymus (Figure 5D), in the red pulp of the spleen (Figure 5E), and in the epithelium of the bursa of Fabricius (Figure 5F). The IFITM3 mRNA levels in 20-day-old yellow-feathered broilers were also investigated with RT–PCR and SYBR–PCR. IFITM3 mRNA was expressed in the three immune organs (Figure 5G,H), with the highest expression in the bursa.
IFITM3 expression in ARV-infected CEF
cells and yellow-feathered broilers
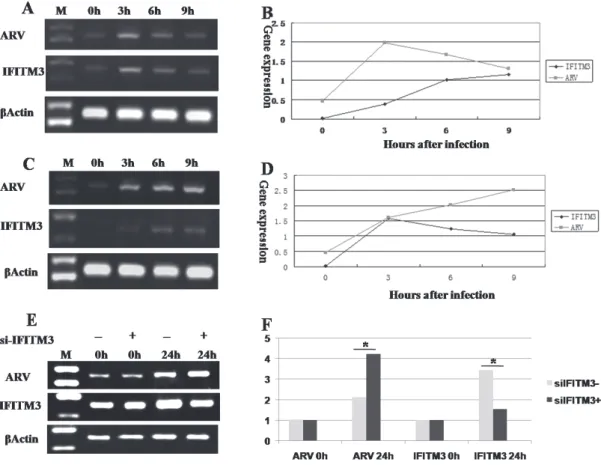
After ARV infection 12h and 24h, the mRNA expression levels of IFN-α and IFITM3 in CEF cells was higher than the control group (Figure 6 A,B).
To investigate the function of IFITM3 after ARV infection, the mRNA expression levels of IFITM3 and IFN-α in the thymus, spleen, and bursa were determined with RT–PCR and SYBR-qPCR. IFN-α expression was
induced by ARV, and then stimulated the expression of IFITM3 early in infection. IFITM3 expression in the thymus (Figure 6 C, D) and bursa (Figure 6 E, F) peaked on day 3 after ARV infection, and then gradually decreased. In the spleen (Figure 6 G, H), IFITM3 began to increase on day 1 after infection, and peaked on day 2. Therefore, ARV stimulated the secretion of IFITM3 in the early stage of ARV infection.
IFN induces IFITM3 expression to inhibit
ARV replication
Cultured CEFs were infected with ARV. IFN-α (20 ng/ mL) was added to the culture fluid of the experimental group and an equal amount of physiological saline was added to the control group. In CEFs with IFN activity, ARV expression peaked at 3 h, and then gradually decreased, whereas the expression of FITM3
continued to increase gradually (Figure 7 A, B). But ARV expression gradually increased in IFN-null CEFs, and that the expression of IFITM3 peaked at 3 h, gradually decreasing thereafter (Figure 7 C, D).
RNA interference downregulates the
expression of IFITM3 and affects ARV
replication
CEF cells were transfected with siRNA of IFITM3 for 0 or 24 h. The RNA was then extracted and reverse transcribed to cDNA, using β-actin as the internal control. RT–PCR was used to determine the effects of
the different periods of IFITM3 interference on ARV expression. When interference at 0 h was compared with no interference at 0 h, neither IFITM3 nor ARV expression changed. After interference for 24 h,
IFITM3 expression decreased relative to that in the interference-free control, showing that a low level of RNA interference was achieved (Figure7 E, F). ARV expression increased when IFITM3 expression was inhibited with IFITM3 interference. With no interference, ARV expression decreased. These results indicate that IFITM3 protein could inhibitreplication of ARV.
Figure 7 – Parte A e B: In CEFs with IFN activity, ARV expression peaked at 3 h, and then gradually decreased, whereas the sion of FITM3 continued to increase gradually. Parte C e D: ARV expression gradually increased in IFN-null CEFs, and that the expres-sion of IFITM3 peaked at 3 h, gradually decreasing thereafter. Parte E e F: After interference for 24 h, IFITM3 expresexpres-sion decreased relative to that in the interference-free control, showing that a low level of RNA interference was achieved.
DISCUSSION
There are five members of the IFITM family in humans, IFITM1, IFITM2, IFITM3, IFITM5, and IFITM10, and these proteins are all induced by type I and type II IFNs (Laura et al., 2014). They are involved in many physiological activities, including cell adhesion, cell differentiation, cell signal transduction, antitumor and antiviral activities, and immune surveillance (Perreira et al., 2013). Research recently showed that IFITM was an important antiviral protein, resisting many kinds of viral
infections. The replication of influenza virus, West Nile virus, and Dengue virus was significantly inhibited by
The IFN-induced expression of the IFITM proteins is ubiquitous. Although IFITM5 is only expressed in bone cells, other IFITM-encoding genes are widely expressed in tissues and organs (Siegrist et al., 2011). IFITM3 is mainly distributed on or in the cell membrane, and it inhibits viral entry into the cell (Bailey et al., 2012). The immune organs (thymus, spleen, and bursa) of birds play important roles in resisting viral infection. Our experimental results found that the IFITM3 gene was expressed in the immune organs of chickens, suggesting that IFITM3 was associated with the chicken’s immune function.
Does IFITM3 suppress the proliferation of chicken viruses? Smith et al. found that the IFITM3 protein of chickens inhibited the replication of ARV (Smith et al., 2013). ARV is one of the most important viruses in poultry production, and understanding the effect of
IFITM3 on ARV infection is important for the discovery of new strategies to impede ARV transmission. In this study, we found that ARV activated the expression of IFN-α and IFITM3 in the immune organs (thymus, spleen, and bursa of Fabricius) in 20-day-old yellow-feathered broiler chickens artificially infected with ARV. The expression of IFITM3 was also induced in ARV-infected CEFs. When IFITM3 was strongly expressed after its induction by IFN, the replication of ARV was inhibited, but when the IFITM3 gene was knocked down with shRNA, the replication of ARV was enhanced. Therefore, we consider that the FITM3 protein could inhibit the replication of ARV.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENT
This work was supported by the Chicken Industry System Program of Fujian China (NO. K83139297) and the National Spark key Program of China (2015GA720001).
COMPLIANCE WITH ETHICAL
STANDARDS
All chicks used in the experiments were treated in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of China. The animal protocol used in this study was approved by the Research Ethics Committee of the College of Animal Science, Fujian Agriculture and Forestry University, Fujian, China.
REFERENCES
Anafu AA, Bowen CH, Chin CR, Brass AL, Holm GH. Interferon-inducible transmembrane protein 3 (IFITM3) restricts reovirus cell entry. The journal of biological chemistry 2013;288(24): 7261–17271.
Bailey CC, Huang IC, Kam C, Farzan M. IFITM3 limits the severity of acute influenza in mice. PLoS Pathog 2012;8:e90991002.
Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. IFITM proteins mediate the innate immune response to influenza A H1N1 virus, West Nile virus and Dengue virus. Cell
2009;139(7): 1243–1254.
Canoui E, Noël N, Lécuroux C, Boufassa F, Sáez-Cirión A, Bourgeois C, Lambotte O. IFITM1expression in Cd4 T cells in HIV controllers is correlated with immune activation. Jaids-Journal of Acquired Immune Deficiency Syndromes 2016; Aug 19. [Epub ahead of print] PMID: 27552157.
Chen YS, Shen PC, Su BS, Liu T., Lin CC, Lee LH. Avian reovirus replication in mononuclear phagocytes in chicken footpad and spleen after footpad inoculation. Canada Journal Veterinary Research 2015;79(2): 87–94. Chen YX, Welte K, Gebhard DH. Induction of T cell aggregation by antibody
to a 16kd human leukocyte surface antigen Immunology 1984;133: 2496–2501.
Lu J, Pan Q, Rong L, He W, Liu SL, Liang C. The IFITM proteins inhibit HIV- 1 infection. Journal of virology 2011; 85( 5) : 2126-2137.
Lorena V, Irene L, José M, Javier B. Avian reovirus sigmaA localizes to the nucleolus and enters the nucleus by a nonclassical energy-and carrier-independent pathway. Journal of virology 2009; 83(19): 10163– 10175.
Miller LC, Jiang Z, Sang Y, Harhay GP, Lager KM. Evolutionary characterization of pig interferon-inducible transmembrane gene family and member expression dynamics in tracheobronchial lymph nodes of pigs infected with swine respiratory disease viruses. Veterinary Immunology and Immunopathology 2014;159(3-4):180-191.
Perreira JM, Chin CR, Feeley EM, Brass A.L. IFITMs restrict the replication of multiple pathogenic viruses. Journal of Molecular Biology 2013;425(24):4937-55.
Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signaling, antiviral responses and virus countermeasures.
Journal of Gene Virology 2008;89:1–47.
Smith SE, Gibson MS, Wash RS, Ferrara F, Wright E, Temperton N, Kellam P, Fife M. Chicken interferon-inducible transmembrane protein 3 restricts influenza viruses and lyssaviruses in vitro. Journal of Virology 2013; 87(23):12956-12966.
Siegrist F., Ebeling M, Certa U. The small interferon-induced transmembrane genes and proteins. Interferon Cytokine Reseach 2011; 31:183-197. Teng LQ, Xie ZX, Xie LJ, Liu JB, Pang YS, Deng XW, Xie ZQ, Fan Q, Luo SS.
Complete genome sequences of an avian orthoreovirus isolated from Guangxi, China. Genome Announc 2013;1(4): e00495-13.
Wang QX, Wu YJ, Cai YL, Zhuang YB, Xu LH, Wu BC, Zhang YD. Spleen transcriptome profile of Muscovy ducklings in response to infection with Muscovy duck reovirus. Avian Disease 2015;59(2):282-290. Yount JS, Moltedo B,Yang YY. Palmitoylome profiling reveals
S-palmitoylation-dependent antiviral activity of IFITM3. Nature Chemestry Biology 2010; 6:610-614.
Multiplication by Ifitm3 published in the Revista Brasileira de Ciência Avícolas/Brazilian Journal of Poultry Science, v20 (2):305-316, in page 380, 381, 382 and 383 where it was written:
Figura
the correct form is Figure
http://dx.doi.org/10.1590/1806-9061-2017-0662
Figura
the correct form is Figure