Identifying Stable Reference Genes for qRT-PCR Normalisation in Gene Expression Studies of Narrow-Leafed Lupin (Lupinus angustifolius L.).
Texto
Imagem




Documentos relacionados
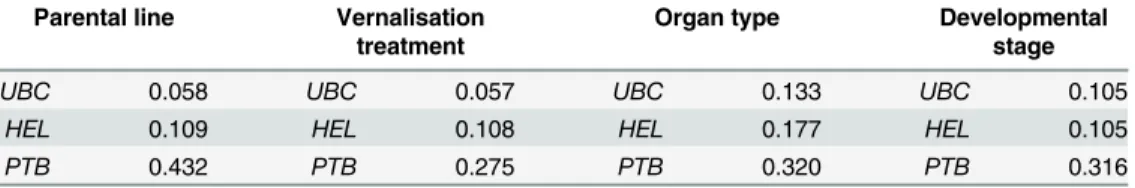
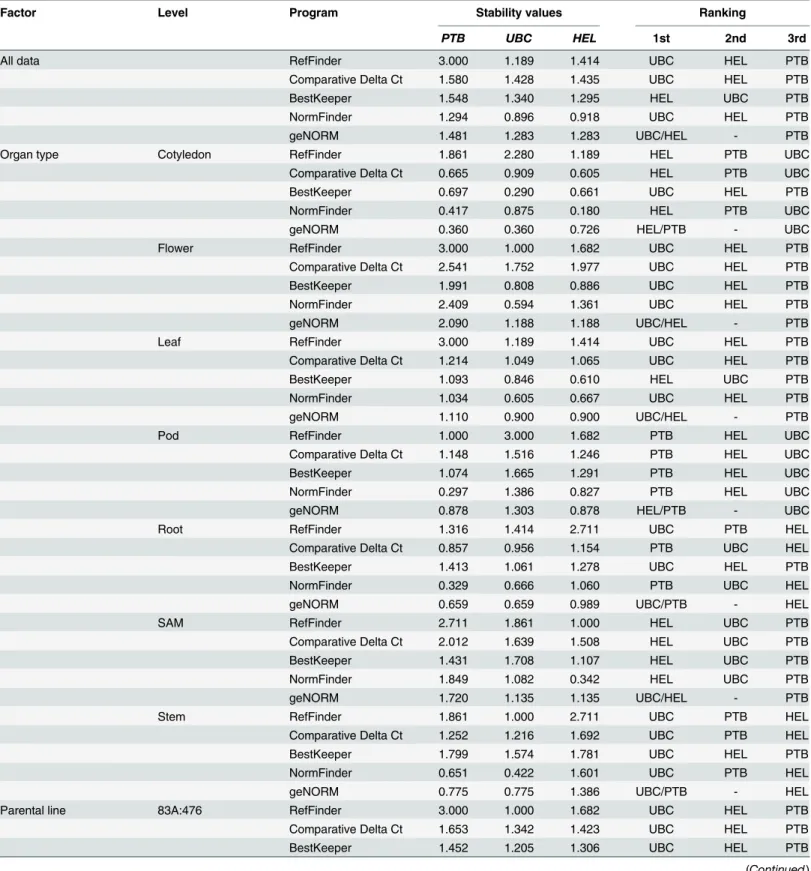
Taken all together, our results confirmed that the most stable reference genes ( GAPDH and ACTIN ) identified in current study could be used for the normalization of gene
The expressions of most genes display variable expression levels in prenatal and postnatal periods, and reference genes that are frequently used for other experiments are not
To identify the most suitable reference genes for RT-qPCR analysis of target gene expression in the hepatopancreas of crucian carp (Carassius auratus) under various conditions
This study aimed at evaluating the stability of candidate reference genes in order to identify the most appropriate genes for the normalization of the transcription in rice and red
To distinguish the effect of sleep deprivation on gene expression stability, M values were also calculated for each group (C, PSD96, SR24 and TSD6) independently, but all M values
In this study, we assessed the suitability of six reference genes frequently used in the literature ( ACTB , B2M , GAPDH , HPRT1 , TBP and TFRC ) using samples from 3 sites within
The case of South Korea Oxford Review of Education Waldow, Florian 2017 Projecting images of the ‘good’ and the ‘bad school’: top scorers in educational large-scale assessments as
Analysis of expression stability using geNorm, NormFinder and BestKeeper revealed that UNK2 , PP2A and SAND could be considered to be appropriate reference genes for gene
(2003), regarding sex-dimorphic expression of these genes, the top two ranked reference genes at each time point from this study are used to compare against a selection of three