Molecular variability in the maize grey leaf spot pathogens in Brazil
Kátia R. Brunelli
1, Larry D. Dunkle
2, Cândido A. Sobrinho
3, Ana C. Fazza
1and Luis E.A. Camargo
1 1Departamento de Entomologia, Fitopatologia e Zoologia Agrícola, Escola Superior de Agricultura
“Luiz de Queiroz”, Universidade de São Paulo, Piracicaba, SP, Brazil.
2United States Department of Agriculture, Agricultural Research Service,
Crop Production and Pest Control Research Unit, Purdue University, West Lafayette, Indiana, USA.
3Embrapa Meio Norte, Teresina, PI, Brazil.
Abstract
Isolates ofCercospora species from leaves displaying symptoms of grey leaf spot were collected in maize-producing areas of south-central Brazil in 2001 and 2002. Restriction digests of the internal transcribed spacer region of rDNA detected the presence of the same twoCercospora species described on maize in the United States, namely C. zeae-maydis and the recently described species, C. zeina. Genetic variability among isolates was assessed by ana-lysing 104 amplified fragment length polymorphism loci. Cluster analysis confirmed the genetic separation of isolates into two species with a mean similarity of 35%. Similarity levels within species were high, averaging 93% and 92% among isolates ofC. zeae-maydis and C. zeina, respectively. The mean genetic similarity between C. zeae-maydis andC. zeina and two isolates of C. sorghi f. sp. maydis was 45% and 35%, respectively. Results of this study showed that populations of the grey leaf spot pathogens in Brazil are similar to those in the United States regarding species composition and thatC. zeina is also present in Brazil.
Key words: Cercospora zeae-maydis,Cercospora zeina, epidemiology.
Received: March 18, 2008; Accepted: September 1, 2008.
Maize grey leaf spot disease is caused byCercospora zeae-maydis, butCercospora sorghif. sp.maydisis occa-sionally isolated from infected leaves (Chupp, 1953). Wanget al.(1998) demonstrated that populations of the grey leaf spot pathogen in the U.S. are comprised of two sibling species, which they designated C. zeae-maydis
group I and group II. Despite their morphological similar-ity, the two groups are substantially divergent based on analysis of amplified fragment length polymorphism (AFLP) data. In addition to the distinct AFLP profiles, the two groups can be distinguished by the restriction patterns of the internal transcribed spacer (ITS)-5.8S rDNA region. In comparative analyses of ITS-5.8S rDNA nucleotide se-quences of Cercospora species and anamorphs of
Mycosphaerella, Goodwin et al. (2001) concluded that groups I and II represent different species. Recently, Crous
et al. (2006) applied DNA phylogeny of nucleotide se-quences of five nuclear-encoded genes and analysed mor-phological characteristics of isolates of the grey leaf spot pathogen from South Africa and compared them with iso-lates of the pathogen from the U.S.A. They confirmed the
distinct separation of two species, provided complete de-scriptions, and named the African pathogen Cercospora zeina, replacing the previous designation ofC. zeae-maydis
group II.
In the U.S., populations ofC. zeae-maydis(group I) andC. zeina (group II) are sympatric in portions of the maize-producing regions, althoughC. zeinais restricted to the eastern third of the country (Wanget al., 1998). Signifi-cantly, the African population of the grey leaf spot patho-gen is comprised solely ofC. zeina(Dunkle & Levy, 2000; Okoriet al., 2003), although the possibility of a third maize pathogen more closely aligned withC. apiiandC. beticola
was suggested by Crouset al.(2006). The considerable ge-netic divergence between the species, their differential geo-graphical distribution, and slightly greater haplotype diversity of African isolates, together with historical re-cords, led Dunkle and Levy (2000) to propose thatC. zeina
originated in Africa, although they did not rule out the pos-sibility of independent origins in the U.S. and Africa.
The occurrence of grey leaf spot in Brazil was first re-ported in 1934 (Viégas, 1945), but it remained a disease of secondary importance until 2001, when severe outbreaks were reported in the central states (Fantinet al., 2001). The reasons for this shift in the severity and distribution of the disease are unclear but may be due to the introduction or emergence of more aggressive genotypes of the pathogen www.sbg.org.br
Send correspondence to Luis Eduardo Aranha Camargo. Depar-tamento de Entomologia, Fitopatologia e Zoologia Agrícola, Escola Superior de Agricultura “Luiz de Queiroz”, Av. Pádua Dias 11, 13418-900 Piracicaba, SP, Brazil. E-mail leacamar@esalq.usp.br.
or to changes in maize hybrids or cropping systems or a combination of those factors. Thus, the objective of this study was to assess the genetic diversity of Brazilian iso-lates of the grey leaf spot pathogens collected in 2001/2002. Our results suggest that well-established populations of both species are present in Brazil and that within-species genetic diversity is very low. This is the first report of the occurrence ofC. zeinain Brazil and assessment of the ge-netic variability among isolates of the grey leaf spot patho-gen.
A collection of 69 monoconidial isolates of the grey leaf spot pathogen was obtained by direct isolation from diseased maize leaves randomly collected from production areas in the south, southeast, and west-central regions of Brazil (Table 1). Conidia were removed from the lesions, transferred onto potato dextrose agar (PDA) plates, and in-cubated for 3 to 5 days at 27 °C until typical colonies devel-oped. For DNA extraction, mycelium was obtained from stationary liquid cultures grown in 200 mL of PD (po-tato-dextrose) medium for 14 days under continuous light at 27 °C. The DNA from four American and two African isolates of the grey leaf spot pathogen, previously classified into genetic groups I or II (Wanget al., 1998), and two iso-lates ofC. sorghif. sp.maydiswere included in the analysis for genetic comparative purposes (Table 1).
To evaluate the production of cercosporin, cultures were grown on PDA and incubated at 27 °C for 14 days in constant light. Colonies secreting a reddish-purple pigment into the medium were scored positive for cercosporin pro-duction.
DNA was extracted from approximately 100 mg of freeze-dried mycelium according to Reader and Broda (1985). The isolates were analysed by restriction fragment length polymorphism (RFLP) of the ITS-5.8S rDNA region after digestion with TaqI (New England Biolabs) as de-scribed by Wanget al.(1998). In addition, individual diges-tions withMseI,BfaI,DdeI (New England - Biolab),HhaI, andHaeII (Fermentas) were also tested to investigate the existence of unique restriction patterns that may indicate additional species or genetic variability. The ITS-5.8S rDNA fragment was amplified by PCR with primers ITS4 (5’ TCC TCC GCT TAT TGA TAT GC 3’) and ITS5 (5’ GGA AGT AAA AGT CGT AAC AAG G 3’) (Dunkle and Carson, 1998). The reaction was carried out in a final vol-ume of 50µL according to Milleret al.(1999). Digestions
were performed in a final volume of 20µL with 5µL of the
PCR product and 8 units of each restriction endonuclease. Restriction fragments were separated by electrophoresis in a 3% agarose gel. AFLP amplifications were performed ac-cording to Voset al.(1995) with the same primer combina-tions used by Wang et al. (1998). Genomic DNA was digested with EcoRI and MseI and ligated with double-stranded adaptors. Preamplification was performed with theEcoRI primer adapter plus the selective base A and with the MseI primer adapter plus the selective base A or C.
Table 1- Identification and origin ofCercospora spp.isolates used in this study.
Isolate Origin Isolate Origin
Uber1 Uberlândia, MG Gua2.2 * Guaíra, SP
Uber2 Uberlândia, MG Gua5 Guaíra, SP
Uber3 Uberlândia, MG SJBV São João da Boa Vista, SP
Uber4 Uberlândia, MG Mig1 Miguelópolis, SP
Uber5 Uberlândia, MG Mig2 Miguelópolis, SP
Uber6 Uberlândia, MG Mig3 Miguelópolis, SP
Uber7 Uberlândia, MG Mig4 Miguelópolis, SP
Uber8 Uberlândia, MG Mig5 Miguelópolis, SP
Uber9 Uberlândia, MG Mig6 Miguelópolis, SP
Uber10 Uberlândia, MG Lem Luis Eduardo Ma-galhães, BA
U1.1 Unaí, MG Castro Castro, PR
U1.2 Unaí, MG Per Perolândia, GO
U2.1 Unaí, MG Ja1.3 Jardinópolis, SP
U2.3 Unaí, MG Ja3.1 Jardinópolis, SP
CD2.1 Cachoeira Dourada, MG JB1.2 José Bonifácio, SP
CD2.2 Cachoeira Dourada, MG JB1.3 José Bonifácio, SP
CD2.3 Cachoeira Dourada, MG Pira1.1 Piracicaba, SP
CD3.1 Cachoeira Dourada, MG Pira4 Piracicaba, SP
CD3.2 Cachoeira Dourada, MG Pira5 Piracicaba, SP
CD3.3 Cachoeira Dourada, MG Pira6 Piracicaba, SP
I1 Indianópolis, MG Pira7 Piracicaba, SP
I2.2 Indianópolis, MG Pira7.1 Piracicaba, SP
I2.3 Indianópolis, MG Pira9.3 * Piracicaba, SP
I3 Indianópolis, MG Pira10.1 Piracicaba, SP
I3.2 Indianópolis, MG Pira11.1 Piracicaba, SP
I4.1 Indianópolis, MG Pira11.2 Piracicaba, SP
I4.2 Indianópolis, MG Pira11.3 Piracicaba, SP
I5 Indianópolis, MG Pira12 Piracicaba, SP
I6 Indianópolis, MG RV1 Rio Verde, GO
I7.1 Indianópolis, MG Cris1 Cristalina, GO
I7.2 Indianópolis, MG Cris2 Cristalina, GO
I8 Indianópolis, MG Cris3 Cristalina, GO
I9.1 Indianópolis, MG 34 Iganga, Uganda -Africa
I9.2 Indianópolis, MG 24 Iganga, Uganda -Africa
I10 Indianópolis, MG 19 Oley, Pennsylvania - USA
I11 Indianópolis, MG 15a South Charleston, Ohio - USA
PO Presidente Olegá-rio, MG
14B Sheridan, Illinois -USA
Tup Tupaciguara, MG 11D Sheridan, Illinois -USA
Irai4.1 Iraí de Minas, MG
Preamplified DNA fragments were diluted 1:5 (v/v) with ultra pure water and used as templates for the selective am-plifications with primerEcoRI + A and eitherMseI + AT or
MseI + CA. PCR products were separated by electrophore-sis at 80W and a maximum temperature of 50 °C for 4 h in a 6% polyacrylamide gel in 1X TBE buffer. The gel was stained with silver nitrate and developed in sodium carbon-ate (Cresteet al., 2001). Enzyme digestion and AFLP am-plifications were carried out twice with a subset of eleven isolates in order to evaluate the reproducibility of the AFLP profiles.
For each isolate, AFLP loci were visually scored as present or absent and the AFLP profile was converted into binary data. Genetic similarity between isolates was calcu-lated using the Dice coefficient based on paired compari-sons of AFLP profiles, as described by Levyet al.(1993). A phenetic cluster (dendrogram) was obtained by the un-weighted pair-group method with arithmetic averaging (UPGMA). Analyses were performed with the NTSYSpc version 1.8 statistical software (Exeter Software, Setauket, NY), and the dendrogram was constructed with the MEGA II - version 2.1 software program (Kumaret al., 2001).
The species composition of the grey leaf spot patho-gen population was determined by restriction analysis of the PCR-amplified ITS-rDNA region (Wanget al., 1998). Digestion of the 520 bp long fragment amplified from all isolates withTaqI generated three different restriction pat-terns, which exactly match the patterns (not shown) ofC. zeae-maydis,C. zeina, andC. sorghidescribed by Wanget al. (1998). In all, 41 isolates were classified as C. zeae-maydis(group I type), 28 asC. zeina(group II type), and two asC. sorghif. sp.maydis. As expected, the control African isolates 5-34 and 5-24, and the U.S. isolate 19 ex-hibited the expectedC. zeinarestriction pattern, whereas the three other U.S. isolates (1-5, 14B, and 11B) exhibited theC. zeae-maydispattern. Of the other five restriction en-zymes tested, onlyBfaI failed to digest the ITS fragment;
HhaI produced an identical RFLP pattern for all isolates;
MseI distinguished isolates ofC. zeae-maydisfrom isolates ofC. zeinabut did not distinguishC. zeinafromC. sorghif. sp. maydis, whereas restriction patterns from HaeII and
DdeI digestions distinguishedC. zeae-maydisandC. zeina, but did not distinguishC. zeae-maydisfromC. sorghif. sp.
maydis. Isolates ofC. zeae-maydisandC. zeinawere de-tected in samples from all states, except Goiás, where only
C. zeinaisolates were detected.
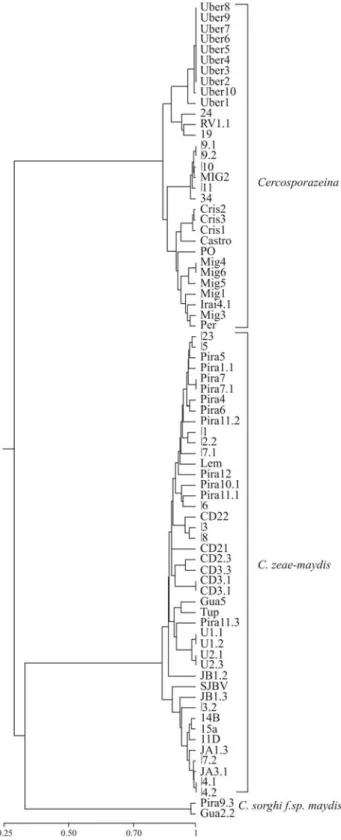
The genetic variability among isolates comprising each species was determined by AFLP analysis. Both primer combinations generated over 150 AFLP loci, but for comparative analyses only those fragments ranging from 50 to 350 bp, a total of 104 AFLP loci, were included in the analysis. These scored loci were also present in the repli-cate AFLP amplifications performed with a sub-set of eleven isolates. Isolates grouped into two clusters, with a mean genetic similarity of 35% between them (Figure 1).
respectively. The mean similarity amongC. zeae-maydis
isolates from Brazil and the United States was 90%, whereas amongC. zeinaisolates the mean similarity was 90.2%. The mean similarity between African and Brazilian isolates ofC. zeinawas 89%. A third group, consisting of the two C. sorghif. sp. maydis isolates, displayed mean similarities of 45% and 35% with isolates of C. zeae-maydisandC. zeina, respectively.
All 69 isolates grew well on PDA, producing greenish grey mycelium. The growth rate ofC. zeae-maydisisolates was consistently and noticeably faster than that ofC. zeina. Of the 41 isolates ofC. zeae-maydisanalysed, 26 (63%) produced cercosporin, but none of the 28 isolates of C. zeinaproduced the toxin.
The AFLP analyses and the two distinct ITS-5.8S rDNA restriction patterns indicated the presence of two di-vergent species that correspond to those reported to occur in the U.S. Isolates of both species were recovered from samples taken from locations separated by as far as 1,500 km. The widespread occurrence of such highly diver-gent groups suggests that the grey leaf spot epidemics in Brazil were caused by factors other than a sudden change in the genetic composition in the population of the grey leaf spot pathogen, or by a recent introduction of more aggres-sive genotypes. Among the possible factors are the rela-tively recent modifications in maize cultivation in Brazil, which include the widespread use of susceptible hybrids, the extension of the total cropping season resulting from an increase in acreage of late-season crops notably in the west-central regions, and the adoption of the reduced-tillage cropping system known to favour the survival of the pathogen (Payneet al., 1987; de Nazarenoet al., 1993). To-gether, these practices may have contributed to a build-up of inoculum that led to epidemic. Thus, it appears that the history of occurrence of grey leaf spot in Brazil is similar to that in the U.S., where the pathogen was not regarded as economically important for several years after it was first reported, but became a major pathogen concomitant with incorporation of changes in cultivation (Roaneet al., 1974; Latterell and Rossi, 1983; Wardet al., 1999).
The within-species levels of genetic similaritiy among Brazilian isolates of both species were nearly identi-cal to those reported for U.S. isolates: 93 and 92% for Bra-zilian isolates compared to 93 and 94% for U.S. isolates of
C. zeae-maydis andC. zeina, respectively (Wang et al., 1998; Dunkle and Levy, 2000). However, the variability betweenC. zeae-maydisandC. zeinawas slightly higher in the U.S., where the species were separated by a mean ge-netic distance of 80%, compared to Brazil, where the mean genetic distance was 65%. Although both studies used the same AFLP primer combinations and analysed a similar number of loci (111 in the U.S. study and 104 in this study), the discrepancy may reflect a sampling effect of the spe-cific AFLP loci scored and of the number of isolates ana-lysed. Among the 91 isolates included in the U.S. study,
only 12 wereC. zeinaisolates compared to 28 of the 69 Brazilian isolates. Nevertheless, the extent of genetic vari-ability observed within and between Brazilian species sup-ports the conclusions of Wanget al.(1998) that the two groups they defined in the U.S. represent distinct gene pools and are likely to be different species, as suggested by Goodwinet al.(2001) and recently confirmed by Crouset al.(2006).
This study also demonstrated a high genetic similarity among Brazilian, U.S., and African isolates. The mean sim-ilarity between Brazilian and U.S. isolates of C. zeae-maydis was 90%, a value comparable to the variability found within this species in both countries, as discussed above. Brazilian isolates ofC. zeinawere on average 90.2% and 89% genetically similar to the U.S. and African isolates of the same species, respectively. Similarly, the East Afri-can population ofC. zeina was found to lack population structure and was indistinguishable from the U.S.C. zeina
(group II) population based on AFLP and RFLP analyses (Okoriet al., 2003). Thus, there appears to be limited ge-netic variability within populations ofC. zeinaworldwide, which is consistent with the likelihood that this species re-produces asexually and indicates that geographically sepa-rated populations do not differ substantially from the founding population, which is proposed to be in Africa (Dunkle and Levy, 2000; Crouset al.2006).
Nevertheless, the most obvious and consistent fea-tures distinguishing the two species are the faster growth and production of the reddish-pigmented phytotoxin cer-cosporin only by isolates ofC. zeae-maydis.
Grey leaf spot can be managed by the use of geneti-cally resistant hybrids. Thus, inferences on the durability of resistance genes are of great interest to plant breeders. De-spite significant interactions between host genotypes and environments regarding resistance to Cercospora zeae-maydis (Thompson et al., 1987; Bubeck et al., 1993; Lehmensieket al., 2001), the low genetic variability within and between species of the grey leaf spot pathogen assessed from populations from three very distinct geographical ar-eas (U.S.A., Africa, and Brazil) suggests that variability in disease severity is related to environmental factors rather than to genetic variation in the pathogen. Given the relative genetic uniformity of the respective asexual pathogen pop-ulations, the sources of quantitative resistance in maize germplasm will most likely remain effective in Brazil, U.S.A., and Africa.
References
Bubeck DM, Goodman MM, Beavis WD and Grant D (1993) Quantitative trait loci controlling resistance to gray leaf spot in maize. Crop Sci 33:838-847.
Chupp C (1953) A Monograph of the Fungus GenusCercospora. The Ronald Press, New York, 667 pp.
Creste S, Tulmann Neto A and Figueira A (2001) Detection of sin-gle sequence repeat polymorphisms in denaturing polyacryl-amide sequencing gels by silver staining. Plant Mol Biol Rep 19:299-306.
Crous PW, Groenewald JZ, Groenewald M, Caldwell P, Braun U and Harrington TC (2006) Species ofCercosporaassociated with grey leaf spot of maize. Stud Mycol 55:189-197. de Nazareno NRX, Lipps PE and Madden LV (1993) Effect of
levels of corn residue on the epidemiology of gray leaf spot of corn in Ohio. Plant Dis 77:67-70.
Dunkle LD and Carson ML (1998) Genetic variation in Cercosporaand the potential impact on selecting for resis-tance to gray leaf spot of corn. In: Proc of the 53rd Annual Corn & Sorghum Res Conf. American Seed Trade Associa-tion, Chicago, pp 30-35.
Dunkle LD and Levy M (2000) Genetic relatedness of African and United States populations of Cercospora zeae-maydis. Phytopathology 90:486-490.
Fantin GM, Brunelli KR, Resende IC and Duarte AP (2001) Boletim Técnico IAC - A Mancha de Cercospora do Milho. IAC, Campinas, 19 pp.
Goodwin SB, Zismann V and Dunkle LD (2001) Phylogenetic analysis ofCercosporaandMycosphaerellabased on the in-ternal transcribed spacer region of ribosomal DNA. Phyto-pathology 91:648-658.
Kumar S, Tamura K, Jakobsen IB and Nei M (2001) Mega II: Mo-lecular Evolutionary Genetic Analysis. Bioinformatics 17:1244-1245.
Latterell FM and Rossi A (1983) Gray leaf spot of corn: A disease on the move. Plant Dis 67:842-847.
Lehmensiek A, Esterhuizen AM, van Staden D, Nelson SW and Retief AE (2001) Genetic mapping of gray leaf spot (GLS) resistance in maize. Theor Appl Genet 103:797-803. Levy M, Correa-Victoria FJ, Zeigler RS, Xu S and Hamer JE
(1993) Genetic diversity of the rice blast fungus in a disease nursery in Colombia. Phytopathology 83:1427-1433. Miller RN, Quezado-Soares AM and Lopes CA (1999) Molecular
comparison ofFusariumpopulation causing eumartii wilt and dry rot of potato in Brasil. Fitopatol Bras 24:149-155. Okori P, Fahleson J, Rubaihayo PR, Adipala E and Dixelius C
(2003) Assessment of genetic variation among East African Cercospora zeae-maydis populations. Afr Crop Sci J 11:75-85.
Payne GA, Duncan HE and Adkins CR (1987). Influence of till-age on development of gray leaf spot and number of airborne conidia ofCercospora zeae-maydis. Plant Dis 71:329-332. Reader UE and Broda P (1985) RAPD preparation of DNA from
filamentous fungi. Lett Appl Microbiol 1:17-20.
Roane CW, Harrison RI and Genter CF (1974) Observations on gray leaf spot of maize in Virginia. Plant Dis Rep 58:456-459.
Thompson DL, Bergquist RR, Payne GA, Bowman DT and Goodman MM (1987) Inheritance of resistance to gray leaf spot in maize. Crop Sci 27:243-246.
Viégas AP (1945) Alguns Fungos do Brasil. Boletim 8. Sociedade Brasileira de Agronomia, Rio de Janeiro, 160 pp.
Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M,et al.(1995) AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res 23:4407-4414.
Wang J, Levy M and Dunkle LD (1998) Sibling species of Cercosporaassociated with gray leaf spot of maize. Phyto-pathology 88:1269-1275.
Ward JMJ, Stromberg EL, Nowell DC and Nutter Jr FW (1999) Gray leaf spot - A disease of global importance in maize pro-duction. Plant Dis 83:884-895.
Associate Editor: Everaldo Gonçalves de Barros