F
ACULDADE DEE
NGENHARIA DAU
NIVERSIDADE DOP
ORTOCreation of a database based on
artificial intelligence in order to
understand the role played by biofilms
on outbreaks
Ana Patrícia Pinheiro de Melo
Integrated Master in Bioengineering Supervisor: Idalina Machado Co-Supervisor: Manuel Simões
c
Creation of a database based on artificial intelligence in
order to understand the role played by biofilms on
outbreaks
Ana Patrícia Pinheiro de Melo
Integrated Master in Bioengineering
Resumo
Um surto pode ser definido como a ocorrência generalizada de uma doença numa comunidade num espaço de tempo específico, a uma taxa superior à habitual. A ocorrência de surtos constitui um problema grave de saúde pública, uma vez que estes podem afectar grande parte da população. Atualmente, tanto os meios de comunicação como organizações de saúde, tais como a World Health Organization (WHO) e o Centers for Disease Control and Prevention (CDC), demonstram uma preocupação crescente em consciencializar a população acerca da existência de surtos. Para tal, como meio de informar a população, listas atualizadas acerca dos surtos existentes em todo o mundo são fornecidas através de plataformas online. Estes recursos estão, de facto, em concordân-cia com a preferênconcordân-cia dos utilizadores em relação a recursos em papel, sendo mais completos, convenientes e imediatos.
As bactérias existem na natureza na forma plactónica ou sessil (biofilmes). Biofilmes são aglomerados de células microbianas que desempenham um papel importante tanto na causa de doenças como na manutenção da saúde, podendo estar presentes em ambientes naturais, industriais ou hospitalares, quer em superfícies vivas (bióticas) quer em superficies não vivas (abióticas).
A existência de biofilmes promove a proliferação e a sobrevivência de microrganismos que podem estar na origemm de surtos. Quando vivem em biofilme, as bactérias estão protegidas contra pressões externas, o que podem facilitar a sua sobrevivência, mesmo quando são expostas, por exemplo a tratamentos e desinfeção.
No entanto, a existência de biofilmes é frequentemente desconsiderada quando as causas dos surtos são investigadas e, portanto, as vias de tratamento e medidas preventivas escolhidas po-dem não ser as mais adequadas, considerando que, por exemplo,as bactérias que constituem os biofilmes apresentam uma resistência cerca de 1000 vezes superior à forma planctónica.
Embora grandes quantidades de dados de biofilme estejam já disponíveis, a forma como essas informações são apresentadas não é padronizada sendo, hoje em dia, apresentada sob a forma de arquivos individuais, o que torna a determinação de relações entre conceitos mais difícil. A implementação dos métodos in silico para organização de grandes volumes de dados biológicos apresenta várias vantagens que podem potencializar a descoberta de padrões e tendências, uma vez que estas ferramentas permitem ao utilizador facilmente analisar e fazer previsões num curto espaço de tempo.
Não existe, à data, uma plataforma que reúna informações sobre os surtos e o papel desem-penhado pelos biofilmes na sua ocorrência, sendo difícil estabelecer uma relação de causa-efeito entre ambos. A existência de uma base de dados com este tipo de dados condensados seria muito útil para a decisão do tratamento mais adequado e para atribuir aos biofilmes a sua importância como agentes causadores de surtos. Assim, este trabalho surge da necessidade de responder a esta questão, apresentando uma base de dados completa, nova e fiável, com uma interface onde todas estas questões podem ser respondidas.
ii
Esta dissertação, integrada num projeto de Tese de Mestrado, reporta o processo de desenvovi-mento de BioBreakD, uma base de dados baseada em inteligência artificial que permita compreen-der o papel desempenhado pelos biofilmes nos surtos alimentares e na área da saúde.
Abstract
An outbreak can be defined as a widespread occurrence of a disease in a community at a particular time, in a frequency higher than usual. Outbreaks present a serious public health threat since they can affect a large group of the population.
Nowadays, the media and renowned organizations like the World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC) show an increased preoccupation in enhancing the population awareness and knowledge about outbreaks. As a way to inform the general public, updated lists of the current outbreaks existent worldwide are provided in online platforms. In fact, these online resources match with the user’s preference over printed ones, being complete, convenient and immediate.
Bacteria exist in nature as planktonic or as sessile cells, also known as biofilms. This com-munity of microbial cells play an important role both in causing disease and maintaining health, being present in natural, industrial and hospital settings, both in living and non-living surfaces.
It is a known fact that the existence of biofilms promotes the proliferation and survival of some species that act as the causative biological agents of outbreaks. When living in biofilm communities, bacteria are protected against external pressures, which can support their survival, even when exposed to treatments.
However, the existence of biofilms is very unconsidered as causative agents of outbreaks, and, therefore, the chosen treatment paths and preventive measures might not be the most suitable. This can be due to the fact that, cleaning and disinfection procedures do not consider biofilm presence, and that bacteria within biofilms present a resistance that can be 1000 higher than in the planktonic form.
Although large amounts of biofilm data is available, the way that these important information is presented is non-standardized and in individual files, which makes the establishment of relation-ships between concepts hardly possible. The implementation of in silico methods present several advantages that can potentiate the finding of patterns and tendencies in large amounts of biological data, since these tools allow the user to analyze and make fast predictions for large sets of data.
To the author’s knowledge, there is no platform that gathers all the information about out-breaks and the role played by biofilms on their occurrence, being hard to establish a relationship between both. The existence of a DB with this type of condensed data, (rather than dispersed in literature), can be useful for treatment decision and to assign to biofilms their importance as outbreak causative agents. Thus, this work emerged to answer the needing to develop a complete, new and reliable database, with a user interface where all these questions can be answered.
This dissertation, integrated on a Master’s Thesis project, reports the road to the development of BioBreakDB, a database based on artificial intelligence in order to understand the role played by biofilms in food and health outbreaks.
Agradecimentos
Aos meus orientadores, Dra. Idalina Machado e Prof. Manuel Simões, por toda a partilha de conhecimento, paciência e apoio durante a realização desta dissertação.
À Faculdade de Engenharia da Universidade do Porto, por me dar uma casa durante estes 5 anos longe da minha terra natal.
Aos meus pais e ao meu irmão, pelo carinho e apoio durante todo o meu percurso académico e por todos os sacrifícios que fizeram para que este fosse possível.
À Nobe, por seres muito mais que a Madrinha que eu pedi que fosses. Por me acompanhares sempre e me ajudares a tentar ser melhor, não só na Tuna mas na vida. Por seres um exemplo de trabalho, dedicação e de persistência e por nunca teres deixado de acreditar naquilo que sabias que eu era capaz. Pelas sessões de riso e de choro e, sobretudo, por nunca me deixares sentir sozinha. À Rafa, por seres quem és. Por seres a minha companhia. Por todos os projectos em que te irritaste comigo. Por todas as manhãs em que eu adormeci. Por todos os momentos em que cobriste por mim porque eu estava na Tuna. E porque, apesar de tudo isto, nunca questionaste a nossa amizade. Obrigada por aceitares toda a minha maluqueira e estupidez e por seres uma constante nos meus dias.
À Zéni, por teres sido um porto seguro durante estes 5 anos. Acompanhaste-me desde o primeiro dia e, honestamente, sem ti não teria aqui chegado. Obrigada por todas as sebentas e apontamentos, por todas as explicações que não tinhas de dar e por toda a dedicação. Obrigada por todas os jantares de gordice e chazinhos da procrastinação, todas as noitadas na ST (sempre com boa banda sonora) e por estares por perto, mesmo quando estás longe.
Ao Rufus, por tudo o que aprendeste, só para me poderes ensinar. Obrigada por me mostrares a importância do Google. Obrigada por todos os cofis, sopas e passeios até Química. Obrigada por seres um Padrinhão à maneira.
Às minhas Pipocas, Eulália e Frisbee, por acreditarem em mim. Por todas as demonstrações de confiança e de afecto. Por todos os pequenos momentos em que me fizeram sentir especial. Obrigada por todas as dores de cabeça, berros e choradeiras. Obrigada por fazerem de mim uma Madrinha cheia de orgulho e me fazerem querer ser o melhor exemplo possível.
À Su, Ju, Sagres e Carlinha, por nunca me abandonarem, mesmo quando a distância entre nós aumentou. Por me darem mais uma razão para ter saudades de casa e por todos os cafés com conversa aleatória. Que venham muitos mais anos juntas e nunca se esqueçam, We are fabulous.
Aos meus Católicos da FEUP e Bichas Veganas, Blibunda, TONEcas, Sardo e Renatinho, pela amizade, convívio e por me aturarem, que eu bem sei que não é fácil. Obrigada pela partilha de conhecimentos e de memórias e por serem caras amigas numa multidão de desconhecidos.
Por fim, à Tuna Feminina de Engenharia Da Universidade do Porto, pelas amizades, pelos momentos, pelas alegrias, pelas oportunidades e pelo crescimento. Irá comigo pela vida fora o amor à camisola e o orgulho imenso de fazer parte desta Família.
Patrícia
"Eu sei, Que o tempo não pára, Tempo é coisa rara E a gente só repara, Quando ele já passou"
Miguel Gameiro
Contents
1 Introduction 1 1.1 Context . . . 1 1.2 Motivation . . . 1 1.3 Objectives . . . 2 1.4 Structure . . . 2 2 Biological Background 5 2.1 Outbreaks . . . 5 2.1.1 Outbreak characteristics . . . 5 2.1.2 Outbreak investigation . . . 6 2.1.3 Outbreak treatment . . . 8 2.2 Biofilms . . . 8 2.2.1 Biofilm observation . . . 102.2.2 Biofilm structure and phenotype . . . 10
2.2.3 Biofilm properties . . . 12
2.2.4 Biofilm formation . . . 14
2.3 Outbreak and Biofilm relation . . . 15
2.3.1 Cholera . . . 15 3 In Silico Background 19 3.1 In SilicoScience . . . 19 3.2 Databases . . . 20 3.2.1 Introduction . . . 20 3.2.2 phpPgAdmin . . . 21 3.2.3 PostgreSQL . . . 22
3.3 Scientific Paper Selection and Harvesting . . . 22
3.3.1 Search Engine Choice . . . 22
3.3.2 Article Search Approach . . . 23
3.3.3 Web Scraping . . . 25
3.4 Text Mining . . . 26
3.5 Natural Language Processing . . . 26
3.5.1 spaCy and ScispaCy . . . 29
3.5.2 Module Choice . . . 30
3.5.3 Abbreviation Detector . . . 31
3.5.4 Named Entity Recognition . . . 31
3.5.5 Noun Chunks . . . 32
3.5.6 Entity Linking . . . 32
3.5.7 Rule-based Matching . . . 33 ix
x CONTENTS
3.6 State-of-the-art . . . 33
3.6.1 Scientific databases . . . 33
3.6.2 Scientific text mining . . . 35
4 Database and Data Collection 37 4.1 Scientific Paper Harvesting Results . . . 37
4.2 Database Architecture . . . 41
4.3 Data Extraction Process . . . 43
4.4 Database Feeding . . . 46
5 Web Platform 49 5.1 Methods . . . 49
5.1.1 HTML . . . 49
5.1.2 PHP . . . 50
5.2 Proposed Architecture and Functionalities . . . 51
5.2.1 Home . . . 51 5.2.2 Explore Database . . . 51 5.2.3 Statistics . . . 51 5.2.4 Learn . . . 51 5.2.5 More . . . 52 5.2.6 Admin Features . . . 52
5.3 BioBreakDB - Implementation and Prototype . . . 52
6 Conclusions and Future Work 61 6.1 Future Work . . . 62
A 63
B 65
List of Figures
2.1 Steps existent in an outbreak investigation . . . 7
2.2 Different medical environments where biofilm presence is usual . . . 9
2.3 CSLM and SEM biofilm visualization . . . 10
2.4 Illustration of biofilm common features . . . 11
2.5 Schematic representation of biofilm formation steps . . . 14
3.1 Number of publications on all topics by year . . . 20
3.2 Relational and non-relational databases structure . . . 21
3.3 phpPgAdmin user platform for database visualization and manipulation . . . 21
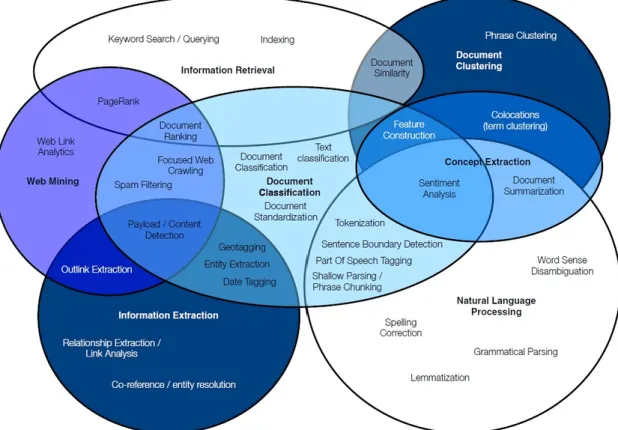
3.4 Inter-relationship among the several TM techniques and their principal function-alities . . . 27
3.5 Overview of the NLP pipeline . . . 28
3.6 Example of the morphological analysis of the word Unexpected . . . 28
4.1 Steps of the implementation of the AI based feeding of the database. . . 37
4.2 Paper selection algorithm . . . 40
4.3 Architecture of database developed . . . 42
4.4 Strain extraction algorithm. . . 44
4.5 Number of cases extraction algorithm. . . 44
4.6 Number of deaths extraction algorithm. . . 45
4.7 Database feeding algorithm. . . 46
4.8 Interconnection between the platform and database tools . . . 47
5.1 Perceived benefits of online platforms . . . 49
5.2 BioBreakDB Home page presentation . . . 52
5.3 Display of BioBreakDB services on Home page . . . 53
5.4 Presentation of the DB data on Explore Database page . . . 53
5.5 Result of the filtering of the DB data with the filters "Disease = Legionaires’ Dis-ease", "Minimum Cases = 50" and "Country = United States of America" . . . . 54
5.6 Presentation of an outbreak page verified by the BioBreakDB administrator . . . 54
5.7 Presentation of an complete outbreak page, not verified by the BioBreakDB admin 55 5.8 Graphical representation of the number of cases and deaths of the diseases out-breaks and the number of outout-breaks reported over time . . . 55
5.9 BioBreakDB Learn page presentation . . . 56
5.10 List of free reviews from Scopus and PubMed indexed with "biofilm" and "outbreak" 57 5.11 Map of concepts extracted with en_core_sci_md UmlsEntityLinker . . . 57
5.12 Data suggestion form from BioBreakDB More page . . . 58
5.13 Contact form, Log in form and Download data option available on More page . . 58
5.14 Example of email receive by the administrator after contact form submission . . . 59 xi
xii LIST OF FIGURES
5.15 List of user suggestions on Admin More page . . . 59 5.16 Admin data verification options . . . 59 A.1 Location correction algorithm . . . 63 B.1 Rule Matchers used for number of cases and number of deaths extraction . . . . 65
List of Tables
2.1 Biofilm composition . . . 12
2.2 General features and advantages of microbial growth as a biofilm . . . 13
3.1 ScispaCy models, training dataset, entity types and F1-score of NER. . . 30
3.2 Labels existent for each NER of the models used in this dissertation . . . 31
3.3 Output from en_core_sci_md model’s UmlsEntityLinker to the entity "Biofilm" . 33 4.1 Number of results of the query ’biofilm[MeSH Terms] AND outbreak[MeSH Terms]’ on PubMed and ’KEY("outbreak" AND "biofilm")’ on Scopus with different fil-ters applied. The search was performed on 08/09/2020. . . 38
4.2 Result of the query ’outbreak[MeSH Terms] AND cholera[MeSH Terms]’ in PubMed and ’KEY("outbreak" AND "cholera")’ in Scopus with different filters applied. The search was performed on 09/09/2020. . . 39
4.3 Number of results of the restriction of having at least one year and one city in the title or abstract of the papers resultant from the filters performed on Table4.2. GPE label refers to countries, cities and states. . . 40
Abbreviations
AI Artificial IntelligenceAPI Application Programming Interface ASP Antibiotic-Surviving Population
CDC Centers for Disease Control and Prevention CSLM Confocal Scanning Laser Microscopy CUI Concept Unique Identifier
DB Database
DBMS Database Management System DOI Digital Object Identifier EPS Exopolysaccharide
HTML HyperText Markup Language HTTP HyperText Transfer Protocol NER Named Entity Recognition NL Natural Language
NLM National Library of Medicine NLP Natural Language Processing NLU Natural Language Understanding MBC Minimum Bactericidal Concentration
MBEC Minimum Biofilm Eradication Concentration MeSH Medical Subject Headings
PHP Hypertext Preprocessor POS Part Of Speech
REGEX Regular Expression
RDBMS Relational Dabatase Management System SEM Scanning Electron Microscope
SQL Structured Query Language TM Text Mining
TUI Type Unique Identifier
UMLS Unified Medical Language System URL Uniform Resource Locators WHO World Health Organization WWW World Wide Web
Chapter 1
Introduction
1.1
Context
Bacteria exist in nature as planktonic cells, free-living bacteria, or as sessile cells, also known as biofilms. According to the National Institutes of Health (NIH), among all microbial and chronic infections, 65% and 80%, respectively, are associated with the presence of biofilms [1].
The existence of biofilms promotes the proliferation and survival of the species that cause outbreaks, even when exposed to treatments. Bacteria, when living in biofilm communities, are highly protected against external environmental pressures, like chemical treatments. When condi-tions are favorable, or when biofilms are mature, bacteria tend to slough-off in form of aggregates, colonizing new surfaces. This phenomena promotes bacteria proliferation and survival which can be, many times the cause of outbreaks.
The Centers for Disease Control and Prevention (CDC) defines outbreak as “the occurrence of more cases of a disease than would normally be expected in a specific place or group of people over a given period of time”1. Outbreaks present a serious public health issue, causing the death of millions. For instance, some of the worst pandemics in History include the Russian Third cholera pandemic of 1839–1856 and the Honk Kong Flu of 1968-1969, which are estimated to have deci-mate over a million people each [2,3]. Nowadays, we are dealing with the Novel Coronavirus, an outbreak of respiratory illness first detected in Wuhan, China. This pandemic has, at the date of September 6th, 2020, nearly 27 million cases confirmed and 900 000 deaths [4].
1.2
Motivation
The existence of biofilms is very unconsidered when the causes of outbreaks are investigated and when the most suitable treatments to combat outbreaks are chosen as well as when preventive measures are planned. Almost all antimicrobial and immunological tests are routinely developed using planktonic cultures, that present different phenotypes and properties when compared with
1https://www.cdc.gov/reproductivehealth/data_stats/glossary.html
2 Introduction
bacteria that cause outbreaks [5]. This may lead to inappropriate and ineffective treatment choices, that might promote outbreak proliferation.
On the other hand, the continuous use of the same or similar disinfectant in processing reusable materials supports the selection of resistant clonal lineages, which can lead to the establishment of resident homogeneous bacterial communities in such surfaces. Bacteria in biofilms are then able to adapt to the used disinfectant and survive the reprocessing procedure [6].
Although large amounts of biofilm data is available, it is presented in non-standardized indi-vidual files and, thus, data interchange among researchers and the establishment of relationships between concepts is hardly possible or even attempted [7].
Despite the increasing number of scientific databases (DB), there are other technologies that allow the analysis of the data and that can extract useful information of them, being this analysis of extreme scientific potential.
To the author’s knowledge, there is no platform that reunites all the information about out-breaks and the role of biofilms on their occurrence, which would allow to establish a relationship between both. The existence of DB with biofilm/outbreaks data condensed in one place, (instead of dispersed in literature), would be very useful for treatment decision. Thus, this work merged to answer the needing to develop a complete, new and reliable one.
1.3
Objectives
The main goal of this work is to develop a database, with the use of artificial intelligence based methods (namely Text Mining and Natural Language Processing techniques), to provide the user with information regarding the role played by the existence of biofilms when analyzing cases of disease outbreaks.
This database of outbreak information aims to be easily available to the user, by its pres-ence on BioBreakDB, the web platform (https://paginas.fe.up.pt/~up201505068/ BioBreakDB/home.php) presented on this dissertation.
1.4
Structure
The remainder of this work is structured as follow: Chapter 2presents a biological background, with the state-of-the-art of outbreaks and biofilms and the demonstration of the relation between Cholera outbreaks and biofilm presence; In Chapter3it is explained the in silico background, with the enumeration and description of the computational methods implemented throughout the elab-oration of the dissertation, being also presented the state-of-the-art of existent scientific databases and text mining, as well as their importance and characteristics; Chapter 4, the steps regarding data collection and database feeding are displayed, as well as the results from the web scraping performed on PubMed and Scopus platforms; In Chapter5the importance behind the elaboration
1.4 Structure 3
of a web platform is presented, as well as the list of proposed features and BioBreakDB web-site prototype; At last, in Chapter 6 the conclusions regarding the work of this dissertation are presented, as well as the future work.
Chapter 2
Biological Background
In this chapter, the basic concepts regarding the biological background of this work will be ex-posed. Initially, an introduction on outbreaks will be provided in Section2.1, which will include the concept, as well as the principal characteristics and the outbreak investigation process. Section 2.2, presents information about biofilms, regarding the concept, properties and formation meth-ods. Lastly, in Section2.3 is presented, as a representative example, the relation existent behind the existence of biofilms and Cholera outbreaks.
2.1
Outbreaks
According to the Centers for Disease Control and Prevention (CDC), an outbreak is "the occur-rence of more cases of a disease than would normally be expected in a specific place or group of people over a given period of time"1, as they consider this term a synonym for epidemic (the use of the term outbreak is preferred when making public communications, in order to prevent population panic). It may affect a small and localized group or impact upon thousands of people across an entire continent and can have diverse origins, such as infectious agents and chemical or physical conditions [8]. When an outbreak occurs over a very wide area, affecting a large number of people, it is considered a pandemic1.
2.1.1 Outbreak characteristics
Understanding the characteristics of an outbreak is extremely important in order to be able to control the spreading, treat the ill and prevent its reappearance. Some of the most important aspects to know about an outbreak are the causative biological agent, source and route of transmission. These might already be known when the outbreak is detected but, if not, it is important to get fast information about those characteristics, in order to be able to provide the best treatment for the existent victims and prevent additional ones.
Finding the causative biological agent that is responsible for an outbreak has direct implica-tion in the choice of the most suitable approach to treat the sick, being also helpful to the process
1https://www.cdc.gov/reproductivehealth/data_stats/glossary.html
6 Biological Background
of discovering the source and route of transmission associated, once there are environmental lim-itations for each biological agent. In order to discover it, the clinical symptoms of the infected patients are used as a way to narrow down the range of possible agents, consequently reducing the amount of laboratory work needed to identify the correct biological agent for the outbreak [9]. Outbreaks that have origin in microorganisms are also very problematic since these organisms have the capacity to transfer from a wide range of reservoirs (such as animals, water or soil), being also able to pass from one animal species to another, which increases radically the rate at which the outbreak spreads.
Regarding the source of the outbreak, it is important to find the starting point of the path that leads the biological agent into the victim’s organism. The correct identification of the outbreak source allows its isolation, preventing further contamination and disease spreading [9].
Lastly, the route of transmission is related to the path that connects the source to the affected individuals. This outbreak characteristic is used in order to better understand the people that might have been in contact with the biological agent, potential victims that might benefit from early diagnosis, as well as to find strategic points that can be used to apply disease control measures [9]. Overall, it is safe to say that there is not a fixed number of patients affected necessary in order to a contamination upgrades to an outbreak, being this value variable according to the outbreak origin. For example, according to the CDC, in the case of foodborne outbreaks, it is only necessary that two persons get the same illness from the same contaminated food or drink2.
The boundary that is established for the classification of contamination as an outbreak relies on its definition, and consists of the background rate of the disease in the affected population in a defined temporal period. This background value will result from the realization of a set of activities, that aim to confirm the observed cases, define the outbreak scope, both geographical and temporally, and find cases that will provide information of those affected when compared to epidemiological studies or hospital records [10].
Another outbreak aspect that can increase the difficulty of their discovery and treatment is the fact that some agents can affect a patient that only manifests symptoms after a long time, existing a long period of transmission of the infectious agent in which the patient isn’t aware of his infection. Prime examples of this condition are the human immunodeficiency virus and bovine spongiform encephalopathy, commonly known as mad cow disease [9].
2.1.2 Outbreak investigation
The possibility of outbreak existence can be brought to a healthcare professional attention in sev-eral ways, such as the analysis of individual cases of the disease, a report presented by a healthcare provider (like a hospital or a nursing home), or even through a directed call from the patient to the health department [8].
A proper outbreak investigation is very important and has two main goals: to control and pre-vent the spread of disease, so that the number of people affected in the outbreak doesn’t increase,
2https://www.cdc.gov/foodsafety/outbreaks/investigating-outbreaks/size-extent. html
2.1 Outbreaks 7
and to learn how to prevent similar outbreaks to happen in the future, by understanding why and how they happened in the past. Besides, the education of the general population is also extremely valuable, to raise awareness of the dangers related to the lack of sanitation or the inappropriate and excessive antibiotic use.
The decision to conduct an outbreak investigation is made by the local and state health depart-ments, based on their professional judgment, always based on health regulations. This procedure can be extremely varied, going from a 10-minute phone call to the implication of a single in-vestigator or researcher, to thousands of inin-vestigators and researchers, or even a multinational investigation. This difference will be based on the information collected, that can support the de-cision of the professional to determine if the outbreak is small and controlled or spreading rapidly and a threat to many individuals [8,9].
Figure 2.1: Steps existent in an outbreak investigation. Source: CDC3
In Figure2.1, it’s shown the procedure steps that follow the decision to conduct an outbreak investigation, that, in practice, might have a slightly different order or even multiple steps might be done simultaneously3.
In the beginning, to confirm the existence of the outbreak some aspects need to be checked. To confirm that it is an outbreak: 1) the reported cases of the disease should be proved not to be a misdiagnosis; 2) the number of diagnosed cases should be higher than the number of cases expected for that specific disease and setting; 3) the majority of the patients in the outbreak should have the same infection or similar manifestations [8]. In order to do so, the reported cases are reviewed closely, directly with the patient or indirectly with its medical records. This step has additional importance in cases where a new disease seems to be emerging or when there is a discrepancy between the clinical and laboratory results [10].
Nowadays, is notary the preoccupation from news media and renowned organizations like the World Health Organization (WHO) and CDC to enhance the population awareness and knowledge about outbreaks, existing even online platforms of the referred organizations that provide updated
8 Biological Background
lists of the current outbreaks existent worldwide45. This is an expedite and accessible measure, to keep the population informed, being more prone to report if they have similar symptoms to the ones known of a current disease outbreak.
2.1.3 Outbreak treatment
After the confirmation of the existence of an outbreak, it is important to choose the most suitable treatment available, being so mostly based on the causative biological agent. Since a big part of the outbreaks is the result of a microorganism that causes an infection on the victim’s body, the most common method to treat these outbreaks is the use of antibiotics. Although this treatment is very accessible and effective, the appropriate use of antibiotics is extremely important, being an aggressive short-course treatment the most suitable, since bacterial exposure to antibiotics for long periods of time and/or low doses may induce bacteria resistance [11].
2.2
Biofilms
For a long time, microorganisms were viewed as simple individual cells, suspended in a culture when in favorable conditions or as dormant organisms or spores when environmentally stressed. Even though this hypothesis was very useful to characterize and study bacteria, this assumption is not accurate when analyzing bacteria in their natural state [12].
The concept of biofilms is not new, being these complex structures directly observed in the 17th century when Antoni van Leeuwenhoek examined the “scuff” from his teeth. Since that time, it has become clear that microorganisms prefer to grow in surface-associated communities, defined as biofilms [12,13,14].
There has been an extensive evolution in the original concept of biofilm, which is now de-fined as a sessile microbial consortium with specific characteristics. Biofilms are now accepted as a collection of cells irreversibly attached, either to each other, an interface or a substratum, that is held together by a self-produced matrix of extracellular polymeric substances with altered phenotype regarding their growth rate and gene transcription [15]. The bacterial capacity to form biofilms gave them an evolutionary advantage since these organisms acquire the capacity to form very complex and highly structured multicellular communities [14]. If within biofilms, microor-ganisms are more resistant to harsh environments, since they grow in complex communities, with multiple resistant phenotypes and with a matrix as a barrier that offers protection [12].
Biofilms are known to play an important role both in causing disease and maintaining health, being present in natural, industrial and hospital settings, both in living and non-living surfaces.
4https://www.who.int/csr/don/en/
2.2 Biofilms 9
These structures are essential to support commensal microflora6, that may be helpful when pre-venting pathogen infectivity, by inhibiting the colonization and establishment of pathogenic mi-croorganisms. On the other hand, biofilms may lead to many medical conditions and prolonged infections, colonization in implants and problems regarding dental plaque formation. Profession-als in the medical area need to be always alert for the possibility of biofilm formation and existence due to its implications, such as bacterial persistence and resistance to antimicrobial treatments, be-ing the presence of these structures also considered an additional risk factor for post-operation complications [12].
With this in mind, it is extremely important to acknowledge the possibility of biofilm presence, understanding their structure, characteristics and biological processes, in order to better identify in which cases it may be a risk to people’s health and the most suitable way to eliminate it. In Figure 2.2are presented several medical-related locations that have been identified as potential biofilm harbors. These environments are the result of the industrialization and modern-day lifestyles, being places that men have direct contact with, which leads to an increasing preoccupation for the possible biofilm formation [12].
Figure 2.2: Different medical environments where biofilm presence is usual. Adapted from: [12]
6Microorganisms which are present on body surfaces covered by epithelial cells and are exposed to the external environment (gastrointestinal and respiratory tract, vagina, skin, etc.) [16]
10 Biological Background
2.2.1 Biofilm observation
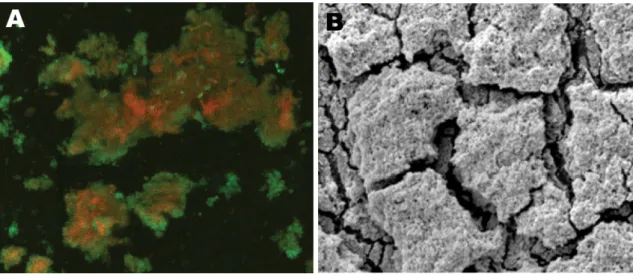
Nowadays, there are several experimental techniques available to harvest accurate information regarding bacterial cell numbers, mode of growth, species composition, and viability, both in planktonic and surface-associated populations. The development of diverse technologies and lab-oratory techniques enables the observation and study of microbial biofilms. Direct observation of biofilms, for example, was facilitated by the application of confocal scanning laser microscopy (CSLM), Figure2.3A, by the development of optically favorable flow cells, as well as by the use of specific probes that allow the determination of species identity and viability [14].
When analyzing the bacterial population in natural ecosystems, direct observation is the gold standard for bacterial enumeration. However, CSLM came to revolutionize this field, since it allows the counting of bacteria on opaque surfaces such as plastics and tissues. This improve-ment of bacterial counting is extremely important since the direct observation occurs without any manipulation, providing solid and unequivocal data of in vivo biofilms [14]. Another important technique that allows biofilm visualization is the scanning electron microscope (SEM), Figure 2.3 B. This method permits the examination and analysis of the microstructure morphology and chemical composition [17].
Figure 2.3: CLSM and SEM biofilm visualization: A) CSLM image of central venous catheter tip in a patient with Nocardia nova complex central line–associated bloodstream infection (original magnification x25); B) SEM image of central venous catheter tip reveals biofilm surface structure (original magnification x5,000). Source: [18]
2.2.2 Biofilm structure and phenotype
Biofilms can show a variety of structures and architectures, that alter with a multitude of envi-ronmental conditions such as temperature and humidity or physical conditions, like pH, charge and surface properties, and also physiological conditions of their microbial content. The heteroge-neous environments that are created within the biofilms also aid in the growth and survival of the
2.2 Biofilms 11
microorganisms since they are able to arrange themselves in the best way to optimize nutrient ex-change and microenvironment stability. Nevertheless, there are some common features to biofilm structure that are important to understand, being very useful for biofilm identification [12,19].
The primary common features of biofilms are: a substratum to bacteria attachment; a con-ditioning film; a biofilm matrix and a liquid or gas phase. These structures are represented in Figure2.4.
Figure 2.4: Illustration of biofilm common features. Source: [12]
The substratum is the interface to which the microorganisms attach in the process of biofilm formation and it can be of abiotic or biotic nature. Regarding abiotic material, they can be, for example, plastics like the ones found in catheters and shunts, titanium metal alloys or ceramics of implants, or even hydrogels like the ones used for contact lenses. Biofilm substratum of biotic origin includes tissue cell surfaces, colonized by bacterial biofilms [12].
The conditioning film is a layer that is naturally formed in any kind of surface and its nature depends both on the properties of the substrate and the chemical composition of the liquid medium. This layer is usually composed of glycoproteins and lipids, such as, for instance, the proteins from urine existent on catheter surfaces, that create a conditioning film where bacteria have the facility to adhere. This layer will also define which bacterial strains will adhere to the substratum first, acting as primary colonizers [12].
One of the most important parts of the biofilm is its matrix. Biofilm bacterial cells are em-bedded in a translucent matrix that fills the 3 to 6 micrometers spaces between cells. This ma-trix also constrains the Brownian movement7 of the bacterial cells [14]. The biofilm matrix is mainly composed of water, proteins, extracellular DNA, ions and exopolysaccharide (EPS) (Table 2.1) [12,20,21].
7movement caused by the molecules in a liquid striking/hitting against the particles of an object leading to shaking and vibration
12 Biological Background
Component Percentage of matrix
Water Up to 97 %
Microbial cells 2-5 % Exopolysacharides 1-2 %
Proteins <1-2 % (includes enzymes) DNA and RNA <1-2 %
Ions Bound and free
Table 2.1: Biofilm composition. Adapted from: [20]
Of the biofilm matrix components, EPS has a very important role, being its composition im-portant not only for adhesion and stabilization of the matrix but also for generating heterogeneity and increasing nutrient availability within the biofilm. EPS is responsible for creating the three-dimensional structure of the biofilm, which is only possible due to this highly hydrated biopolymer that has the capacity of immobilizing bacteria. Matrix EPS is also responsible for the specialized niches within the biofilm and, its production differs according to the environmental conditions as well as the bacteria species that are present within the biofilm, as well as the variety and proportion of different strains. In high-nutrient environments, EPS production increases as well as cell pro-duction, which leads to the existence of denser biofilms. On the other hand, if there is a nutrient deficiency, the biofilm matrix is less dense and inter-dispersed with water channels [12].
2.2.3 Biofilm properties
When bacteria growth within a biofilm, they acquire several advantageous properties, when com-paring to planktonic bacteria. These features are described on Table2.2and are related to protec-tion, nutrient acquisiprotec-tion, new traits and intercellular communication.
The biofilm structure and phenotype can increase bacterial individual cells protection from ex-treme conditions and substances in the environment, working as a physical barrier to the exposure to immunogenic epitopes and to the immune response. Cells growing within biofilms are usually 1000 times more resistant to antimicrobial agents than the planktonic cells. Furthermore, the fact that microbial cells within a biofilm present a slower growth rate and are phenotypically different from planktonic cells, leads bacteria to experience nutrient gradients and waste by-products (due to the different microenvironments existent in the biofilm matrix) that may generate resistance and slower growths. Furthermore, slow growth rates are associated with increased resistance to some antimicrobial compounds. The fact that the biofilm matrix is composed mainly of water also in-creases cell protection against rapid desiccation and, EPS and other adhesins that may exist to hold the matrix form, also help to immobilize bacteria against severe hydrodynamic and host clearance mechanisms [12].
As mentioned before, the biofilm structure improves bacteria nutrient acquisition since it has water channels, that are able to transport soluble nutrients from the bulk fluid to the inner parts of
2.2 Biofilms 13
Feature Description
Protection From host defenses and predators From antimicrobial agents - slow growth rate
- poor penetration - altered phenotype From desiccation
From fluid hydrodynamic and mechanical forces Nutrient
acquisition
Elevated concentrations of nutrients - surface phenomenon
- nutrient trapping
Microbial and environmental heterogeneity for metabolic cooperation Spatial heterogeneity to optimize transport of by-products and increase nutrient influx
New traits Phenotypic plasticity - novel gene expression and bacterial phenotype Plasmid or genetic transfer between organisms
Mutation due to selection Intercellular
communication Quorum sensing/ density-dependent communication Interspecies communication
Table 2.2: General features and advantages of microbial growth as a biofilm. Adapted from: [12]
the biofilm and, similarly, can transport metabolites and waste products out of the biofilm matrix. In biofilms of higher densities, there are not open water channels like in monoculture biofilms but there are less dense regions with a very dynamic population of microorganisms cooperating in community, sharing nutrients and preventing the build-up of toxic by-products [12].
Concerning phenotypic variation, the heterogeneous microbial populations that are within biofilms are able to adapt to new conditions by exhibiting different phenotypes, different from planktonic cells that grow in homogeneous environments. This phenotypic plasticity that can be the result of gene mutations or the expression of different phenotypes due to biofilm heterogeneous growth conditions, resulting in multiple expressions of traits essential for survival [12].
Lastly, intercellular communication occurs when bacteria are able to demonstrate the same behavior under the same environmental conditions. An example of this communication is cell-density-dependent signaling, first observed in Vibrio fischeri when the bacteria would become flu-orescent when a critical mass was reached by the population. This behavior, also called Quorum-sensing signaling, is important in coordinating multicellular behavior in bacteria, regulating sev-eral physiological processes [12].
14 Biological Background
2.2.4 Biofilm formation
The study of a wide variety of rivers and streams made clear the preponderance (superior to 99,99%) of the existence of biofilm cells in these ecosystems and also that their presence is shown to be proportionately active in nutrient cycling. Therefore, biofilms are considered the main mode of bacteria growth in water streams. After the development of biofilm visualization techniques like light and electron microscopy, ecologists reported the presence of biofilms in basically every natural environment, from tropical leaves to desert boulders [14].
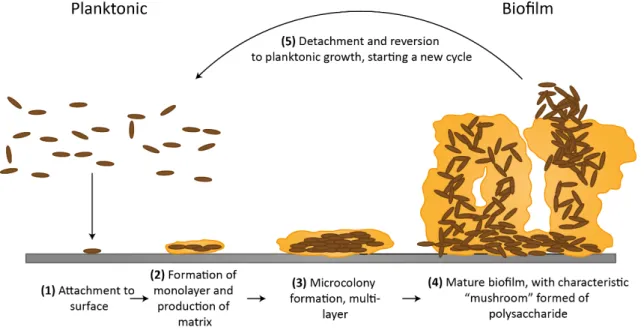
The study of biofilm behavior in water systems is very important since it can help to under-stand the formation and control of naturally occurring processes like corrosion and fouling that frequently provoke several costly problems. For this is crucial to understand the sequence of events that allow biofilm formation, represented in Figure2.5[14].
Figure 2.5: Schematic representation of biofilm formation steps: 1) attachment of planktonic cells; 2) monolayer formation and matrix production; 3) microcolony formation; 4) biofilm maturation; 5) cell detachment. Source: British Society for Immunology8
The dynamic sequence of events that lead to biofilm formation is usually divided in four stages: bacterial attachment as planktonic cells, monolayer formation, microcolony formation and mature biofilm formation. In the end, bacteria detach from the biofilm, in a process termed sloughing-off, returning to the planktonic phase as individual cells or agglomerates that start a new cycle of biofilm development.
During the initial bacterial attachment the contact between bacteria and the interface is a con-sequence of passive movement caused by Brownian movement or gravitational forces [12,20]. The monolayer formation happens after the primary reversible attachment to the surface, which
8https://www.immunology.org/public-information/bitesized-immunology/ pathogens-and-disease/biofilms-and-their-role-in
2.3 Outbreak and Biofilm relation 15
still allows bacteria to easily detach and move along the surface. Environmental factors like pH and temperature influence this reversible attachment as well as the nature of the surface, which de-termines the extent of the adhesion [12,20]. Eventually, the bacteria become irreversibly attached to the surface, leading to microcolony formation. This attachment occurs due to specif interac-tion entities, like adhesins, and non-specif ones, including hydrogen bonds, van der Waals forces and hydrophobic interactions. Studies have also shown a direct relation between the class of the fimbria of the bacteria within the biofilm and the irreversible attachment to the surface [12,20]. Lastly, occurs the mature biofilm formation, being the EPS production the most important step that allows irreversible attachment and development of the three-dimensional structure character-istic of a mature biofilm. In this stage, the microbial cells within the biofilm present different characteristics when compared to their planktonic forms. Eventually, bacteria will disperse and detach from the biofilm, returning to the planktonic phase [12,20].
2.3
Outbreak and Biofilm relation
The existence of a relationship between the bacterial biofilm formation and the increase in antibi-otic resistance is documented. This makes the disregard of their existence during the choice of outbreak treatment unreasonable, leaving, therefore, room to improvement.
As a representative example, it was studied the relation of bacterial biofilms with increased resistance and disease outbreaks of cholera. The choice of this disease was based on its tendency to form disease outbreaks and, since it has a substantial number of cases, it has, therefore, a lot of research available about them.
2.3.1 Cholera
Cholera is estimated to have annually 2.8 million cases, of which 91 000 result in death, being spread across 51 endemic countries [22]. It is an acute secretory diarrheal infection, caused by the gram-negative and motile bacterium of serogroup O1 and 0139 [23,24] and its characterized by profuse secretory diarrhea and vomiting that, in the most severe cases, can lead to dehydration and death by hypovolaemic shock [25]. This causative agent can be divided into two biotypes, namely the classical and El Tor, which identification is extremely important to determine the source and spread of infection [26]. These bacteria are common in aquatic ecosystems, being the risk of outbreak related to the quality of sanitation facilities that the population has access to [22, 24, 25]. Areas with poor sanitation are associated with the rapid dissemination of this disease, taking into advantage the hyperinfective stage of the bacteria that is present in fresh cholera stool. This bacterium can act as a source for new outbreaks since it is more contagious than the V. cholerae in the natural environment, being able to stay in this hyperinfectious state after leaving the human host for hours to days [23].
Vibrio choleraeis known to be able to switch between motile and biofilm lifestyles. Further-more, it is suggested that the biofilm-like structures formed by these bacteria during infection can
16 Biological Background
influence physiology, ecology and epidemiology V. cholerae, being biofilms their preferred mode of survival [24,25,27,28].
Both in the aquatic environment as well as in the human small intestine (bacteria host upon oral ingestion), Vibrio cholerae is exposed to diverse physical, chemical and biological stresses, being the biofilm mode of growth important for protection, giving bacteria higher resistance to these environmental stressors [24,25]. V. cholerae biofilms attach to abiotic and biotic subtracts and the bacteria can be transmitted to humans by ingestion of water with infected copepods (that present a commensal relationship with this bacterium) or from person to person, in cases of poor access to sanitation, via the fecal-oral route, through the ingestion of contaminated water or food [23].
Upon the entrance in the host organism, the bacteria capacity to develop biofilms is critical to intestinal colonization, being biofilm formation initiated quickly after V. cholerae adherence to the intestinal cells. The microscopical analysis shows that V. cholerae biofilm formation starts about 30 minutes after adherence to the intestinal cell line, and the biofilm thickness increases with time [24,28]. After ingestion, it is suggested that the bacteria return to their motile planktonic form to adhere to the host intestinal cells. With the infection progression, V. cholerae assembles once more into biofilm-like structures favored by the chemical conditions (increase resistance to stress caused by changes in pH and the existence of digestive enzymes, p.e.) [28]. Lastly, before exiting the human body, these biofilms with a high concentration of bacteria detach from the intestinal cells (still aggregated) being released in this hyperinfectious form, causing a rapid spread of the disease [28].
Another advantage of early biofilm formation, even before virulence expression, is the in-creased adherence of the bacteria to the host, preventing it to be easily flushed out, in comparison to the planktonic form, that stays in the intestinal lumen. Therefore, it facilitates bacterial colo-nization since prevents its elimination prior to virulence expression [28].
There are also studies that compare the antibiotic resistance between the V. cholerae planktonic and biofilm forms [27,28].
In a recent study of Gupta et al. [27], five different classes of antibiotics were chosen (ampi-cillin, doxycycline, ciprofloxacin, erythromycin and ceftriaxone) and the antibiotic susceptibility was determined with the minimum inhibitory concentration (MIC), minimum bactericidal concen-tration (MBC) and minimum biofilm eradication concenconcen-tration (MBEC) values. The effects of the antibiotic treatment on both forms, as well as the virulence of the antibiotic-surviving population (ASP) were also analyzed. It was found that V. cholerae biofilms showed higher antibiotic resis-tance than its planktonic form (MBEC of biofilm culture higher than MBC of planktonic cultures), for all of the tested antibiotics [27]. Regarding the ASP, it was found that both forms of growth showed the ability to colonize and to produce symptoms upon infection. The ASP clones were also tested in their planktonic and biofilm form and only the biofilm form kept enhanced antibiotic resistance, which indicates that it is exclusive of the biofilm phenotype [27]. Similar results were found in [28], where adhered cells of V. cholerae were less susceptible to all antibiotics tested, namely chloramphenicol, rifampicin, tetracycline and azithromycin.
2.3 Outbreak and Biofilm relation 17
host cells forms biofilms, that facilitate colonization and enhance antibiotic resistance, confirming the danger behind this architectural colony in increasing cholera transmission and facilitating its persistence in water environments, affecting their eradication.
Chapter 3
In Silico Background
In this chapter, the concepts regarding the In Silico background of this work will be presented. Initially, an introduction on In Silico science will be provided in Section3.1. Section3.2, displays information about databases, namely their importance and the computational methods for their development and maintenance. On Section 3.3 is presented the approach behind the scientific paper selection and harvesting, as well as an introduction to the web scraping methods. Section 3.4describes the basics of text mining while section3.5introduces Natural Language Processing and its main functionalities. Lastly, in Section3.6 is presented the state-of-the-art of scientific databases and scientific text mining.
3.1
In Silico Science
In silicomethods refer to methods that use computational approaches to predict results of bio-logical experiments, being the term derived from the computer component silicium. Even though this term was coined in 1987, these methods have gained their importance and popularity more recently, with the technological evolution that made them possible. The need for in silico biol-ogy methods emerged due to the increase of the amount of information available, being these fundamental to apply sophisticated algorithms in order to advance scientific understanding [29].
In silicomethods have several advantages such as the ability to make fast predictions for a large set of data in a high-throughput mode and make predictions based on the structure of a compound without the need for it to be synthesized [29]. Moreover, in silico approaches also help in finding patterns and tendencies in large amounts of biological data.
In a simple approach, in silico methods allow scientists to do science through their computer, without having to spend hours in the laboratory, by figuring out existent relations and patterns, through the analysis and interpretation of complex datasets and published scientific papers. The work proposed for this dissertation fits in this category since it aims to point out the relation between biofilms and outbreak occurrence, through the analysis of published scientific papers.
20 In SilicoBackground
3.2
Databases
3.2.1 Introduction
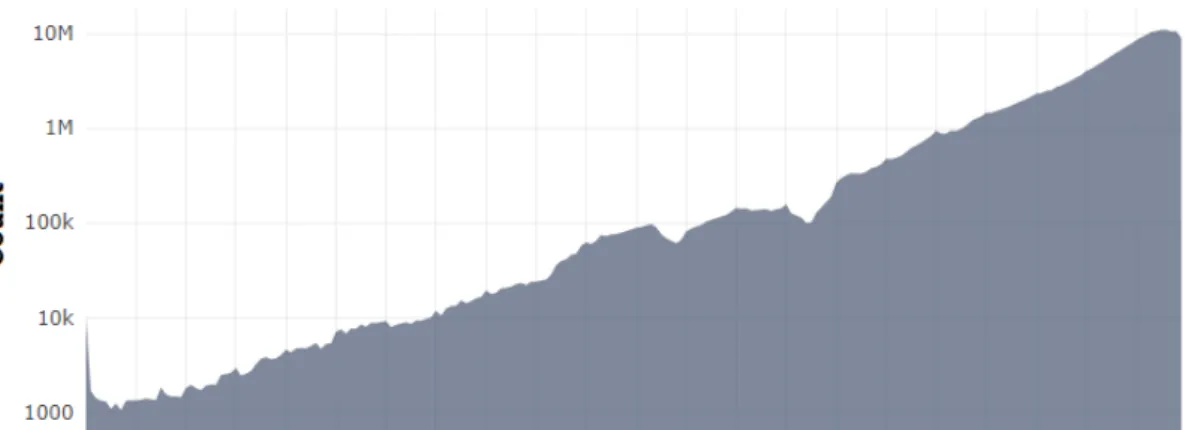
A database (DB) consists of an organized collection of structured information, that is usually stored electronically in a computer system and controlled by a database management system (DBMS)1. DBs can store various types of information, such as store products, details of cus-tomers, or record members of an organization [30]. With the large amounts of data (Figure3.1) that have become available, digital DBs gained importance for data storage and organization, since their appearance in the 60s [31].
As shown in Figure3.1, the number of papers published by year is in constant growth, being the current value for all topics over the 10 million mark. There is an obvious increase in the quantity of information available, justifying, therefore, the use of DBs to manage it.
Figure 3.1: Number of publications on all topics by year. Source: Microsoft Academic2 In the field of bioinformatics, biological DBs play an important role, allowing scientists to access a wide variety of biologically relevant data, such as genomic sequences, that would be very difficult to analyze and store otherwise [32]. Nevertheless, it is suggested in some studies that complementing a DB with links to full texts is needed in order to increase its use and reliability in the scientific field [33].
As shown in Figure 3.2, databases can be divided into two principal groups: relational databases and non-relational databases.
Non-relational DBs don’t use relations (tables) as their storage structures, storing data in a single document instead. This model’s main advantages are that it allows reading and writing data quickly, it supports mass storage, it is easy to expand, and has a low cost. Nevertheless, this type of
1https://www.oracle.com/database/what-is-database.html 2https://academic.microsoft.com/publications/?topLevel=true
3.2 Databases 21
Figure 3.2: Relational and non-relational databases structure. Source: SQLNOSQLDATABASE3 DB doesn’t support Structured Query Language (SQL), which is the industry standard to manage data [34,35].
The relational DB model was invented by E.F Codd in 1970, being its principals described in Codd et al. [36], and soon relational DB management systems (RDBMS) became very popular. This type of DB consists of a collection of data items organized in tables, also known as relations, that can be accessed and reassembled easily. The main components of this relational model are the collections of relations for data storage and a set of operators that can act on relations to produce other relations and data integrity, to ensure accuracy and consistency [34,37].
There are several tools to create and maintain DBs and their tables. In this dissertation, ph-pPgAdmin and PostGreSQL will be used for the development of a relational database, which architecture is presented in Chapter5.
Figure 3.3: phpPgAdmin user platform for database visualization and manipulation
3.2.2 phpPgAdmin
phpPgAdmin is a graphical web-based administration tool for creating and maintaining Post-greSQL databases. It is an open-source software tool, written in Hypertext Preprocessor (PHP), that allows DB administration over the web. Its user-friendly interface allows performance of the
22 In SilicoBackground
most frequently used operations, with intuitive data visualization and manipulation4. As shown in Figure3.3, phpPgAdmin displays on the left side of the page all available schemas, databases and tables, making then easily accessible and, on the right side, it’s shown the wide variety of tools available, as well as the tables or input fields of these tools, that include DB creation, a list of DB restrictions and import of data from files. Moreover, phpPgAdmin has also the ability to directly execute SQL statements. In this dissertation phpPgAdmin 5.1 was used, hosted at db.fe.up.pt5.
3.2.3 PostgreSQL
According to the American National Standards Institute (ANSI) and the International Standards Organization (ISO), which is affiliated with the International Electrotechnical Commission (IEC), the standard language for relational database management systems is SQL6 7.
SQL development has a specific utility - SQL uses a special purpose query language to interact with relational databases [31]. This language is efficient, easy to learn and use and functionally complete, allowing the user to define the relational structure of the DB (data definition language), retrieve and extract information from the relations (data manipulation language) and control the access rights to them (data control language) [37,38] .
There are several free RDBMS available, being the one used PostgreSQL8, due to its compati-bility with phpPgAdmin. It is a free open source software that, since its development in 1996, has gained extreme importance and strong reputation, competing nowadays with the major relational DB vendors such as Oracle and MySQL [38].
PostgreSQL presents multiple advantages that attract both developers and companies. It has no associated licensing costs, is SQL-standards-compliant, gives high performance, is scalable and reliable, being very appealing businesswise. Regarding its user advantages, are to notice its very good documentation and active community, its relative ease to automate DB administrative tasks and its capacity to be integrated with other DBMS [38]. The version running in the host used in this dissertation is PostgreSQL 9.6.19.
3.3
Scientific Paper Selection and Harvesting
3.3.1 Search Engine Choice
According to the need created by the evolution of the electronic age and the exponential growth of available data, several medical DBs were created, available on the Web, enabling the study of specific topics. These include bibliographic databases, where journal articles can normally be found and, therefore, the majority of the scientific research available. The referred DBs usually
4http://phppgadmin.sourceforge.net/doku.php/ 5http://db.fe.up.pt/phppgadmin/
6https://blog.ansi.org/2018/10/sql-standard-iso-iec-9075-2016-ansi-x3-135/ 7https://docs.oracle.com/cd/B13789_01/server.101/b10759/intro002.htm
3.3 Scientific Paper Selection and Harvesting 23
include information regarding the scientific paper (such as title and authors) and links to full-text content, when applicable (open source publications) [39,40].
To select the scientific literature of interest for the development of this dissertation, two of these DBs were used, namely PubMed9 and Scopus10. These, which are amongst the four most popular bibliographic search engines (along with Web of Science and Google Scholar), were cho-sen due to the wide spectrum of information available and their comparable advanced search tools, specifically, keyword search, further discussed in3.3.2[39].
PubMed is a free resource that allows the search and retrieval of literature regarding biomedical and life sciences11. It is a web-portal of the medical database MEDLINE, which is the main bibliographic DB of the United States National Library of Medicine (NLM). It is considered the first interactive searchable DB in the field of medicine (introduced in 1971) and collects the interest of both clinicians and researchers [39,40].
Scopus is an abstract and citation DB, arranged by independent subject matter experts12. When the user is looking for an overview of a certain topic it is the most effective search engine, being the only DB that allows the sorting of the results according to their number of citations. Scopus, was developed in Europe and covers a wider journal range, having, however, its citation analysis limited to articles published after 1995. Unlike PubMed, Scopus DB requires an access fee, since it belongs to a commercial provider, Elsevier13. For this dissertation, nevertheless, a connection to the University of Porto library account was used, allowing the retrieval of information free of charge [39,40].
3.3.2 Article Search Approach
After the selection of search engines, it was necessary to establish an approach for the collection of the scientific papers of interest for the dissertation. The goal of the search was to harvest articles that could support the relation between disease outbreaks and biofilm presence. Being also one of the objectives of this dissertation, the elaboration of a publicly available web platform DB, with outbreak characteristics and information, the articles retrieved should have data regarding a specific outbreak, like an outbreak report. Therefore, the aim was to find outbreak reports that also depicted biofilms.
For PubMed, in order to increase the precision and the efficiency of the MEDLINE DB search, it was chosen to use the NLM’s Medical Subject Headings (MeSH) keywords. These keywords result from the manual indexing of scientific papers according to the discussed topics, arranged in a hierarchy, allowing a fast identification of the articles of interest [41,42,43].
The use of MeSH keywords has several advantages, being a key feature of MEDLINE DB. Since these keywords are selected from a list of official words and phrases, it allows a more effi-cient search since after the establishment of the correct term, the user does not have to be concerned
9https://pubmed.ncbi.nlm.nih.gov/ 10https://www.scopus.com/home.uri
11https://pubmed.ncbi.nlm.nih.gov/about/ 12https://www.elsevier.com/solutions/scopus 13https://www.elsevier.com/
24 In SilicoBackground
with variations of the same concept. The MeSH Browser14is a free tool that facilitates the process of finding the correct MeSH keyword, permitting the search of strings in the controlled vocabulary thesaurus. For example, a MeSH of interest for this dissertation was "Disease Outbreaks" (Unique ID D004196)15, englobing different variations of the concept, such as outbreaks, epidemics and pandemics [42]. Besides that, MeSH searches provide more accurate results, since they assert that the topic of interest is specifically discussed in the paper, instead of only mentioned, retrieving fewer irrelevant citations for the research [42,43]. Moreover, these keywords are available for all PubMed articles unlike the author keywords, that can be unavailable when dealing with articles without open access.
MeSH keywords are exclusive of the MEDLINE database16, which makes the search process for Scopus slightly different. For this platform, the article search is also based on keywords. However, a combination of several keywords must be used, namely, author keywords (assigned to the document by the author), index terms (controlled vocabulary terms assigned to the document, such as MeSH keywords), trade name terms (used to identify a commercial product or service) and chemical names (identifying chemical entities). It is not available a search method that uses only MeSH terms in Scopus, in order to keep the process equal in both platforms. However, the choice of this advanced search field (KEY()) aims to make the selection process as similar as possible, including the MEDLINE MeSH keywords when available.
Furthermore, some additional requirements were established, in order to normalize as much as possible the data retrieved, in order to prevent future fallbacks and improve overall performance. First of all, only articles written in English were considered. Secondly, only papers that have the abstract available openly were used, as well as papers that present a valid digital object identifier (DOI).
The DOI system was presented in 1997 by the Association of American Publishers for im-proving the management and stability of scientific content online [44]. This system assigns an identifier to any online object of intellectual property, which remains unalterable, creating an in-frastructure for the registration and use of digital content17. Since the identifier remains always the same, it is not susceptible to changes of an article’s ownership or location, making it a great long-term option [44]. Furthermore, a link to the article source is easily made, by adding the base URL http://dx.doi.org/. This property was very useful in the development of this DB, since only the DOIs of the articles of interest were extracted being provided after to the user with links to the article on the web [44].
To sum up, the approach for article selection was a keyword search, followed by the obliga-toriness of English language and both abstract and DOI availability. The keywords used for both platforms were “Disease Outbreaks” and “Biofilms”, since we wanted papers that show the rela-tion between biofilms and outbreaks. However, since the initial results were very numerous and disperse, in order to have a bigger control over the output, it was decided to focus on a specific
14https://meshb.nlm.nih.gov/search
15https://meshb.nlm.nih.gov/record/ui?ui=D004196 16https://www.nlm.nih.gov/bsd/medline.html 17https://www.doi.org/index.html
3.3 Scientific Paper Selection and Harvesting 25
disease at a time. Thus, a new keyword was added to the query, correspondent to the disease at study. The results of search on both PubMed and Scopus were then extracted, using a Web scraping process.
3.3.3 Web Scraping
Web scraping, also known as web harvesting or web data extraction, is a process of extracting data from websites, simulating human web surfing. When doing so, the programmer accesses web pages and, through the analysis of the page HyperText Markup Language (HTML) and other files that compose the website, is able to find specific data elements, extract, transform and save them as a structured dataset [45].
This technique, which has gained the interest of the programming community, can be useful in a wide variety of fields of work and provides huge advantages when compared with the tradi-tional web analysis through browsers. Web scrapers are an excellent resource when it is needed a process to quickly gather and process large amounts of data available online. The web scraping program will request the targeted website for resource acquisition and, therefore, data available in the browser will be accessed, stored, and virtually analyzed and presented [46,47,48]. Although some websites can have a design that make difficult the process of scraping information, basically every content existent online can be scraped, only varying the amount of code necessary to do so [49].
In order to implement the described method, Python has been chosen as the programming language, using the Selenium package (Python version 3.8.3). Selenium provides a variety of tools, that aim to enable and support the automation of web browsers18. Therefore, it is possible to simulate actions that a user would do online, through a browser, possibly hidden19, gathering information of interest and saving it on the server’s filesystem, for posterior treatment [48]. After the analysis of the structure of both PubMed and Scopus search and result pages, the data of interest - article’s title, abstract and DOI - were extracted to be subsequently stored in the database. Selenium WebDriver Application Programming Interface (API) uses browser automation APIs to run tests and visualize if everything works as expected, being available for most main browsers such as Firefox, Chrome and Safari. In this dissertation, it was used ChromeDriver version 85.0.4183.87 (compatible with Google Chrome 85)20.
Thus, with the described process, the data of the scientific papers resultant from the search approach described at3.3.2was collected to a dictionary. The dictionaries provident from the two different platforms were compared by DOI, removing duplicated articles. After the proper paper selection, the harvested information was further analyzed, resorting to Text Mining and Natural Language Processing methods.
18https://www.selenium.dev/documentation/en/
19Scopus doesn’t allow to be harvested with a hidden browser due to its security policy against online attacks 20https://chromedriver.chromium.org/

![Figure 2.2: Different medical environments where biofilm presence is usual. Adapted from: [12]](https://thumb-eu.123doks.com/thumbv2/123dok_br/15489848.1041296/29.892.143.789.526.902/figure-different-medical-environments-biofilm-presence-usual-adapted.webp)
![Figure 2.4: Illustration of biofilm common features. Source: [12]](https://thumb-eu.123doks.com/thumbv2/123dok_br/15489848.1041296/31.892.149.785.403.631/figure-illustration-biofilm-common-features-source.webp)
![Table 2.1: Biofilm composition. Adapted from: [20]](https://thumb-eu.123doks.com/thumbv2/123dok_br/15489848.1041296/32.892.261.585.146.306/table-biofilm-composition-adapted-from.webp)
![Table 2.2: General features and advantages of microbial growth as a biofilm. Adapted from: [12]](https://thumb-eu.123doks.com/thumbv2/123dok_br/15489848.1041296/33.892.167.760.140.573/table-general-features-advantages-microbial-growth-biofilm-adapted.webp)
![Figure 3.2: Relational and non-relational databases structure. Source: SQLNOSQLDATABASE 3 DB doesn’t support Structured Query Language (SQL), which is the industry standard to manage data [34, 35].](https://thumb-eu.123doks.com/thumbv2/123dok_br/15489848.1041296/41.892.175.748.152.346/relational-relational-databases-structure-sqlnosqldatabase-structured-language-industry.webp)