Anonymization of clinical data
José Miguel Melo
Mestrado Integrado em Engenharia Informática e Computação Supervisor: Gabriel David
José Miguel Melo
Mestrado Integrado em Engenharia Informática e Computação
Approved in oral examination by the committee:
Chair: Doctor Rui Camacho
External Examiner: Doctor Paulo Oliveira Supervisor: Doctor Gabriel David
Over the past years, with the progress of technology, the amount of data being collected by the IT systems has exponentially grown. By using data mining techniques, this data can be analyzed to find trends and statistics, which are really useful for all companies and industries. The analysis and data sharing for studies became a large industry, with a great impact in all sectors. However, with this comes the concern with individual privacy - there is a huge amount of data which is private and should not be public in any circumstances - so, it is highly needed to find a solution to share and analyse data while protecting privacy. Nevertheless, it is truly important to take into account performance issues as the anonymization process should not hinder the normal functioning of the operational system.
The focus goes to clinical data, which allows medical researchers to learn trends, statistics and relations between certain clinical attributes, such as correlations between gender and a specific disease. These studies and data analysis are very important as they can bring great benefits and knowledge in healthcare. However, maintaining individual privacy is crucial.
To balance the need for rigorous data and the requirement of privacy protection, a study on data anonymization is done and some models and algorithms are discussed. This study allows to propose and develop a practical way to efficiently anonymize clinical data. With this solution, the user can quickly and easily anonymize a given MongoDB dataset, through the provision of a set of configurations. The anonymization is done resorting to well known models and algorithms to protect privacy, associated with specific clinical criteria, restrictions and hierarchies. At the end of the anonymization, an anonymized version of the subset is obtained that meets the selected privacy model, balancing enough privacy versus keeping research value.
The solution is evaluated in terms of performance and some optimizations are proposed and implemented to solve some limitations imposed by the implemented prototype. By using a subset of clinical data that needs to be anonymized, the solution’s applicability is validated for research purposes.
Nos últimos anos, com o avanço da tecnologia, a quantidade de dados armazenados pelos sistemas de informação tem vindo a crescer exponencialmente. Recorrendo a técnicas de data mining, estes dados podem ser analisados para encontrar tendências e estatísticas, que são de grande utilidade para todas as empresas e industrias. Assim, a análise e a partilha de informação para estudos tornou-se uma indústria com grande impacto em todos os setores. No entanto, com isto surgem as preocupações com a privacidade individual - muita informação é privada e não deve ser tornada publica em nenhuma circustância. Portanto, surge a necessidade de encontrar uma solução para partilhar a informação, protegendo a privacidade. Esta solução deve ter em conta problemas de performance para não comprometer o normal funcionamento do sistema.
O foco são os dados clínicos, que permite aos investigadores clínicos encontrar novas tendên-cias, estatísticas e relações entre atributos clínicos, como doenças e género. Estes estudos são de extrema importância por trazerem benefícios e conhecimentos na área da saúde. No entanto, a privacidade individual é crucial.
Para balancear a necessidade de informação rigorosa para contextos de pesquisa e o requisito de privacidade, é feito um estudo sobre anonimização de dados e alguns modelos e algoritmos são apresentados. Este estudo possibilita a proposta e desenvolvimento de uma solução para anoni-mizar dados clinicos de uma forma prática e eficiente. Com esta solução, o utilizador consegue rápida e facilmente anonimizar uma base de dados MongoDB através de uma simples config-uração. A anonimização é feita recorrendo a modelos de privacidade e algoritmos conhecidos, aliados a hierarquias, critérios e restrições clinicas. No final do processo, é obtida uma versão anonimizada da base de dados que cumpre o modelo de privacidade selecionado, balanceando privacidade versus valor de pesquisa.
A solução é avaliada em termos de performance e algumas otimizações são propostas e im-plementadas para resolver algumas limitações impostas pelo protótipo implementado. A aplica-bilidade da solução para contextos de pesquisa é ainda validada utilizando uma base de dados real que contém dados clinicos a ser anonimizados.
First of all, I would like to thank my adviser Gabriel David for the interest shown in the disserta-tion and for all the suggesdisserta-tions given, which showed essential to achieve the best results on this dissertation.
I would also like to express my gratitude to my course colleagues and teachers for all the knowledge that they could provide me during these years as student of Faculdade de Engenharia da Universidade do Porto. This knowledge will be trully important and useful for my future.
I would like to express my gratitude to Glintt, and in particular to my adviser Pedro Rocha, for all the support and help given during the dissertation, which showed essential to accomplish the best results on this dissertation and without which it would be a lot harder to finish it.
In addition, a thank to all my closest friends for being there during all these years, for sup-porting me in all my decisions and for encouraging me to be better and to learn the most I can as student.
Finally, I would like to express a special thank to my family for supporting me during these years, for being there in all good and bad moments and without whom it would be a lot harder to achieve all the goals I achieve during these last years.
1 Introduction 1
1.1 Context . . . 1
1.2 Motivation and Goals . . . 1
1.3 Dissertation Structure . . . 2
2 Anonymize Data for Privacy 5 2.1 Principles . . . 5 2.1.1 Basic Definitions . . . 6 2.2 Privacy Models . . . 6 2.2.1 k-anonymity . . . 7 2.2.2 (δ , k) anonymity . . . 9 2.2.3 kmanonymity . . . 9 2.2.4 Bayes-Optimal Privacy . . . 10 2.2.5 `-diversity . . . 10 2.2.6 t-closeness . . . 12 2.2.7 δ -Presence . . . 12 2.3 Data models . . . 12 2.3.1 Numeric data . . . 13 2.3.2 Categorical data . . . 13 2.3.3 Set-valued data . . . 13 2.4 Approaches to anonymity . . . 13 2.4.1 Generalization . . . 14 2.4.2 Suppression . . . 15 2.4.3 Perturbation . . . 16 2.5 Quality Metrics . . . 17 2.5.1 Discernibility metric (DM) . . . 17 2.5.2 Classification metric (CM) . . . 18 2.5.3 Precision (Prec) . . . 18
2.5.4 Normalized Certainty Penalty (NCP) . . . 18
2.6 Algorithms . . . 19
2.6.1 DataFly . . . 19
2.6.2 Optimal Lattice Anonymization (OLA) . . . 20
2.6.3 Incognito . . . 21
2.6.4 Flash . . . 23
2.7 Tools . . . 24
2.7.1 UTD Anonymization Toolbox . . . 24
2.7.2 ARX - Powerful Data Anonymization Tool . . . 25
2.8.1 Recoding Identifiers . . . 31
2.8.2 Names, Contact information and Identifiers . . . 31
2.8.3 Age and Date of Birth . . . 31
2.8.4 Other Dates . . . 32 2.8.5 Medical dictionaries . . . 32 2.9 Summary . . . 32 3 Solution 33 3.1 Requirements . . . 33 3.1.1 Database . . . 34 3.2 Architecture . . . 34 3.2.1 Anonymization Process . . . 35
3.2.2 Anonymization GUI app . . . 45
3.2.3 Anonymization web service . . . 46
3.3 Anonymization Process Results . . . 48
3.4 Summary . . . 49
4 Optimizations 51 4.1 Streams for Memory Usage . . . 51
4.1.1 Data Processment . . . 51
4.1.2 Save Results to MongoDB . . . 53
4.2 Clustering Pre-processment Phases . . . 54
4.3 Results . . . 58
4.4 Summary . . . 61
5 Results 63 5.1 Information loss . . . 63
5.1.1 Prescribed Medications Collection . . . 63
5.1.2 Consumptions Collections . . . 67
5.2 Performance . . . 70
5.2.1 Privacy Model Config Impact . . . 70
5.2.2 Solution - Initial version . . . 72
5.2.3 Solution - Streaming version . . . 75
5.2.4 Solution - Clusters version . . . 77
5.2.5 Comparison . . . 79
5.3 Validation . . . 80
5.4 Summary . . . 86
6 Web and Desktop Application 89 6.1 Web application . . . 89
6.1.1 View process information loss . . . 91
6.1.2 View useful analytics . . . 91
6.2 Desktop application . . . 92
7 Conclusions and Future Work 95 7.1 Future Work . . . 96
A MongoDB Collections Structure 103 A.0.1 Prescribed Medications . . . 103 A.0.2 Consumptions . . . 106
2.1 Categorical data generalization . . . 13
2.2 Date generalization . . . 14
2.3 Vehicle generalization until suppression . . . 15
2.4 Datafly pseudocode (Source: [Swe02a]) . . . 19
2.5 Lattice of generalizations and generalization strategy (orange trace). . . 21
2.6 Incognito pseudocode (Source: [LDR05]). . . 22
2.7 Lattice evolution for Incognito algorithm (Source: [KPE+12]). . . 22
2.8 Flash algorithm lattice example (Source: [KPE+12]). . . 23
2.9 Outer loop of the Flash algorithm (Source: [KPE+12]. . . 23
2.10 CheckPath(Path, Heap) (Source: [KPE+12] . . . 24
2.11 FindPath(Node) (Source: [KPE+12] . . . 24
2.12 Configuration file, taken from UTD Anonymization Toolbox manual (Source: [KIK16]) 26 2.13 ARX main frame . . . 26
2.14 ARX workflow (Source: [arxc]. . . 27
2.15 ARX hierarchy wizard . . . 28
2.16 (1) Attribute properties configuration. (2) Privacy models selection. (3) Utility metrics and suppression limit. . . 28
2.17 ARX solution exploring . . . 29
2.18 ARX anonymization result analysis . . . 29
2.19 ARX tool API uml (Source: [arxb]) . . . 30
3.1 Anonymization process flow chart . . . 36
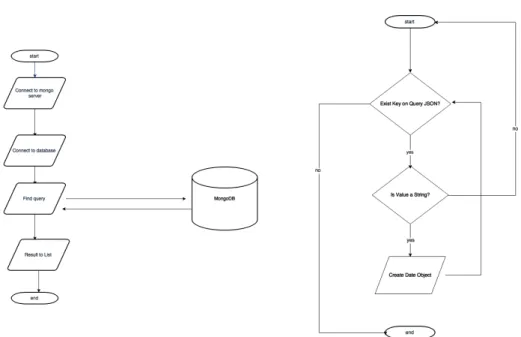
3.2 Connect to mongo server and gather data flowchart (left). Parse dates from query object flowchart (right). . . 38
3.3 Consumptions and Prescribed Medications representations after processment. . . 41
3.4 GUI anonymization process. . . 45
3.5 Web service component diagram. . . 47
3.6 Memory usage for 20k records (left) and memory consumption distribution (right). 49 4.1 Memory usage for 20k (left) and 400k (right) records. [SH] . . . 52
4.2 Memory usage by object for 400k records. [SH] . . . 52
4.3 Clustering architecture. . . 56
4.4 Client-cluster functioning and interaction flowchart. . . 56
5.1 Information loss vs k-anonymity for 20k (left) and 600k (right) records. . . 67
5.2 Memory usage for 20k records (left) and memory consumption distribution (right). [SH] . . . 73
5.3 Memory usage tendency charts. . . 74
6.1 Create new connection (left). Start anonymization (right). . . 90
6.2 Anonymization ended notification. . . 90
6.3 List of results (left). Results for specific anonymization process (right). . . 90
6.4 Information loss view. . . 91
6.5 Web application dashboard with provided analytics. . . 92
6.6 Connect (left) and create the configuration file (right). . . 93
6.7 Create query. . . 93
2.1 Patient’s diseases table . . . 7
2.2 2-anonymity patient’s diseases table . . . 7
2.3 Original glucose table . . . 10
2.4 22-anonymity glucose table . . . 10
2.5 Patient’s diseases table . . . 11
2.6 2-diversity patient’s diseases table . . . 11
2.7 Income table . . . 13
2.8 Anonymized income table . . . 13
2.9 Original table . . . 15
2.10 2-anonymization table with local recoding . . . 15
2.11 2-anonymization table with global recoding . . . 15
2.12 Weight table . . . 16
2.13 Perturbed weight table . . . 16
3.1 Non-hierarchical structure version of Listing3.2. . . 39
3.2 Results for an initial version of the anonymization process . . . 48
4.1 Anonymization process analysis after optimization of data processment and save results phases. . . 59
4.2 Anonymization process analysis after optimization of data processment, save re-sults and validation phases. . . 59
4.3 Anonymization process analysis after clustering architecture implementation. . . 60
5.1 Prescribed Medications anonymization - Information loss for 20k, 100k and 600k records using k=2, k=3 and k=5. . . 66
5.2 Prescribed Medicationsanonymization - Information loss for 20k, 100k and 600k records using `=2, `=3 and `=5. . . 68
5.3 Consumptions anonymization - Information loss for 20k, 100k and 600k records using k=2, k=3 and k=5. . . 70
5.4 Elapsed time for distinct values of k and `. . . 71
5.5 Memory usage for distinct values of k and `. . . 71
5.6 Performance results for the initial version of the anonymization process using k=2. 72 5.7 Performance results for the initial version of the anonymization process using `=2. 73 5.8 Memory usage for 20k records using `=2. [SH] . . . 73
5.9 Memory usage evolution with dataset size. . . 73
5.10 Performance results for the streaming version of the anonymization process using k=2. . . 75
5.11 Memory usage evolution with dataset size. . . 76
5.13 Performance using cluster based solution. . . 77
5.14 Memory usage evolution with dataset size. . . 78
5.15 Memory usage tendency chart for cluster based solution. . . 78
5.16 Elapsed time comparison. . . 79
5.18 Dataset size limitation comparison. . . 79
5.17 Memory usage comparison. . . 80
5.19 Quality metrics returned from anonymization applied to anonymized version of Prescribed Medications dataset. . . 81
EMD Earth Mover’s Distance QID Quasi-Identifier
Prec Precision metric DM Discernibility Metric CM Classification Metric
NCP Normalized Certainty Penalty
HIPAA Health Insurance Portability and Accountability Act RDBMS Relational Database Management System
GUI Graphical User Interface UML Unified Modeling Language EHR Electronic Health Record EMR Electronic Medical Record OLA Optimal Lattice Anonymization JSON JavaScript Object Notation PRMD Privacy Model
Introduction
1.1
Context
The data that is routinely collected in most current organizations can be analyzed and used for research to find trends and statistics. These trends and statistics help researchers and industries to evolve and to find new possible solutions, improvements and support the decision of several layers of Public Administration.
Healthcare is one of the main industries when talking about data sharing for research. The analysis of clinical data, resorting to data mining methods, brings several advantages for the in-dustry and the Health Administration. It helps finding correlations between metrics and attributes, which may lead to new discovers, new possible causes for some diseases, and much other useful information.
However, with all this data sharing comes the concern with individual privacy, which is granted by law and of great importance. So, it is highly needed to find a way of sharing data while preserving privacy. To achieve this, it is required to find ways to anonymize data, that balances privacy and data utility.
From the already said, it is easily understood that this dissertation is focused on database security and privacy, since it addresses ways of anonymizing clinical data to enable data sharing for research purposes.
This dissertation will be developed along with Glintt - Healthcare Solutions. Glintt is one of the biggest technological companies in Portugal and has a focus on healthcare solutions, in which it is a national leader.
1.2
Motivation and Goals
The amount of clinical data is enormous and the analysis of the data collected brings great benefits to all of us. For the pharmaceutical industry, this data is useful as it allows to gain access and
knowledge about how effective some drugs are in the treatment of some diseases, it allows to find out possible secondary effects that were not supposed and possible correlations of these effects with attributes (age, genre, ...), and many other useful information. For clinical research in general, there is an infinite amount of knowledge that can be collected from the analysis of clinical data. For example, it may allow to learn possible causes and correlations of diseases with other factors, the evolution of diseases in our society and why the evolution is happening, or even why a disease is more common in some locations. For our society, this data is extremely important as the analysis of data allows to gain more knowledge and, as consequence, to improve everyone’s healthcare. Final, but not least, it is also important for companies that collect data, as it is an important income source that allows this data to continue being collected and used to study and gather knowledge. However, individual privacy is very important in our society and clinical data can not be provided for research purposes if researchers can identify individuals. Consequently, it is clear the necessity to find the best solution to de-identify individuals in a data set. However, this is not as simple and as linear as it may seem, because there is a huge amount of external information with which clinical data can be associated with to re-identify individuals. This way, it is essential to find the best anonymization method to the problem.
Another important aspect is that, the more encryption and anonymization, the more informa-tion loss. Therefore, it is important to find the optimal soluinforma-tion for an anonymizainforma-tion problem, which is the optimal balance between information loss and amount of anonymization.
Also, data must be always available for medical and clinical entities. So, it is also important to take attention to performance and find a way to not compromise the system while encryption and anonymization process is running.
Electronic Health Records (EHR) include a wide range of data, such as age, diseases, medica-tions, prescripmedica-tions, and many other. So, it is clear that exist many types of data - numeric, names, ... - and it is important that the solution is scalable and flexible enough to handle all these types of data.
The method to solve this problem that was chosen is to create a solution that enables to load a dataset, configure hierarchies for each type of data and identify all required quasi-identifiers, identifiers and sensitive data. After that, the system will process the dataset and return a new anonymized one. At the end of this process, data should be ready for sharing with minimal privacy risk.
1.3
Dissertation Structure
Besides this introduction, this dissertation is divided into 6 more chapters.
In this chapter some concepts and existing work will be covered, which will help in the creation of a solution for clinical data anonymization.
On chapter2some important concepts as well as existing work on the field of anonymization are covered and analysed. Based on this analysis, on chapter3a solution is proposed to solve the problem of clinical data anonymization and a first analysis on an initial version of this solution is
done. In order to improve this initial version, on chapter4some optimizations are analysed and proposed.
With the solution implemented, on chapter5a study on the impact of privacy models in the process of anonymization as well as an analysis and comparison of all implemented versions is done.
On chapter6the web application and the desktop application that resulted from the proposed solution are presented and explained in more detail.
Finally, on chapter7final conclusions on this dissertation are presented and some future work proposed.
Anonymize Data for Privacy
With the growth of technology and data collection, came the necessity of sharing data for research purposes. However, with this, came the concerns about individual privacy and the necessity of anonymizing data.
This topic has been under intense research in the last years in order to try to minimize the risks of breaking individual privacy.
In this chapter some concepts and existing work will be covered, which will help in the creation of a solution for clinical data anonymization.
2.1
Principles
First of all, it is important to clarify the concept of data anonymization. Data anonymization is a process in which a database that contains information about real people is converted into a new database that contains the same information, but in which is not possible to identify any specific person. [Rag13]
Three important characteristics that describe an anonymization problem are privacy model, data modeland quality metrics. [Pod11]
Privacy models are conditions that a database must satisfy to be considered anonymized. The process of data anonymization always implies loss of information. There are several methods that allow to anonymize a dataset but the best method for a problem is the one that less decreases data utility due to information loss. Quality metrics are used to evaluate the quality of the data generated by the anonymization process. These metrics are used to choose the best version of the anonymization process. [Pod11,LGD12].
There are three types of disclosure, which privacy data publishing aims to protect against: [KS+13]
1. attribute disclosure, in which sensitive information is discovered.
3. membership disclosure, in which it is determined if an individual is member of the anonymized dataset.
2.1.1 Basic Definitions
In this section some important definitions will be presented in order to better understand the next sections.
In anonymization problems, and more in the specific case of clinical data anonymization, there are 3 special types of attributes that must be well defined in datasets: sensitive attributes, identifiers and quasi-identifiers. Identifiers, defined at2.1.2, are attributes that identify an individual without the need of external information, such as names. Quasi-identifiers (QID), defined at 2.1.3, are attributes that can identify an individual if external information is available and used. An example of a QID is a diagnosis code, which does not directly identify a patient but if it is a rare diagnosis, then an attacker may infer to whom the diagnosis correspond. A QID can lead to the correct association of a record with an individual, also known as identity disclosure. [BRK+13]
Sensitive attributes, defined at2.1.1, are attributes that are not supposed to be linked to an individual, such as a rare disease. [GDL15]
It is also important to keep in mind the Definition2.1.4of equivalence class as it is an important part of most privacy models that will be covered in Section2.2.
Definition 2.1.1 Sensitive attributes
An attribute that must be kept secret in the anonymized database. [SXF10] Definition 2.1.2 Identifiers
An attribute which explicitly identifies record owner. These attributes are typically excluded from the dataset. [SXF10]
Definition 2.1.3 Quasi-identifier (QID)
An attribute that can identify an individual in the database when combined with external data. [KS+13]
Definition 2.1.4 Equivalence class
Group of rows in which the quasi-identifiers in the selected group of columns have exactly the same values. [EEDI+09]
2.2
Privacy Models
During the last years, with the growth of data and the concerns about individuals privacy, multiple privacy models have been proposed and developed to minimize individual’s privacy risk.
The most well-known models are:
• `-diversity [MKGV07,LLV07] • t-closeness [LLV07]
• o-Presence [NC10]
These models will be analysed and explained in the following sections.
2.2.1 k-anonymity
k-anonymity was introduced by Latanya Sweeney in 2002 to solve the problem of producing a release of data that contains useful data about individuals while those individuals cannot be identified. [Swe,Swe02b].
This privacy model tells that each tuple of QIDs must appear a minimum of k times in order to achieve anonymity [BFW+10,Tho07,AFK+06]
Definition 2.2.1 k-Anonymity
A table is said to achieve k-anonymity if every tuple of quasi-identifiers appear at least k times in that table. [Swe02b]
k-Anonymity is the most common and the most used privacy model because the results are sat-isfactory for most problems and it is the most practical and the easiest to achieve in most scenarios. In order to return good results, quasi-identifiers must be well defined by data publishers.
For better understanding of this model, an example will be shown and explained next. The example is based on the example given by Benjamin C. M. Fung et al. [BFW+10].
Location Sex Age Disease
Porto Male 34 Cancer
Manchester Male 39 Cancer
London Male 38 Diabetes
Porto Male 35 Diabetes
Lisbon Female 24 Hepatitis Lisbon Female 28 Diabetes
Table 2.1: Patient’s diseases table
Location Sex Age Disease
Portugal Male [30-40] Cancer England Male [30-40] Cancer England Male [30-40] Diabetes Portugal Male [30-40] Diabetes Portugal Female [20-30) Hepatitis Portugal Female [20-30) Diabetes Table 2.2: 2-anonymity patient’s diseases table
Example 2.2.1 Suppose Table2.1was intended to be provided for research purposes.
In this table, if an attacker knows that someone lives in Porto and is 34 years old, then the attacker will infer that person has Cancer (row 1).
To prevent this to happen, Table2.1was anonymized with k-Anonymity model, resulting Table 2.2. This new table is 2-Anonymous.
In this anonymized table we notice that every tuple of quasi-identifier - Location, Age and Job - appears at least k times. The tuples are:
• (Portugal, Male, [30-40]) • (Portugal, Female, [20-30)) • (England, Male, [30-40])
If we analyse this table we notice that some generalizations on quasi-identifiers were made: 1. Location - For 2-Anonymity, we needed at least 2 equal values for attribute location, which
is not fulfilled in the initial table due to London and Manchester. So, this quasi-identifier is generalized according to an hierarchy, defined by the data publisher. In this case, it is generalized to Portugal and England.
2. Sex - For 2-Anonymity, we needed at least 2 equal values for attribute sex. This is fulfilled in the initial table, so there is no need to make any changes to this attribute.
3. Age - In the initial table, we had the patient’s exact age. For 2-Anonymity, we needed at least 2 equal values for the age, which is not fulfilled in the initial table. So, this QID is generalized, creating groups: [20,30) and [30,40].
Now, after anonymizing the table, each distinct group of QID appears at least 2 times. So, if an attacker knows that someone lives in Porto and is 34 years old, there will be at least 2 possibilities and so, it is not as simple to find out who has which disease.
To achieve k-Anonymity some approaches are possible. The most well known are: 1. Generalization
2. Suppression
These approaches are better explained in Section2.4.
2.2.1.1 Attacks against k-anonymity
Although k-anonymity is the most common and the most used privacy model, it is vulnerable to attacks, even when k is high and the quasi-identifiers are selected with accuracy and care.
1. Unsorted matching attack - Records are saved into the database sequentially. This way, the order in which the tuples appear on the database can be used as an attack. This type of attack is easily solved by shuffling the anonymized table. [Swe02b]
2. Complementary release attack - Some generalizations of attributes may have the same value or meaning as the initial value, which makes them similar. When this happens, joining the anonymized table with an external information will lead to easily find correlations. Exam-ple: If the quasi-identifier full date is generalized into the year, then the attacker may still find out a relation between the anonymized table and an external table because attributes are similar in both tables. [Swe02b]
3. Homogeneity Attack - Records with similar tuples of QIDs may have the same sensitive attribute.
Example: Suppose an attacker is able to find out which rows in the table may correspond to an individual. Now, suppose one of the attributes in the table is Disease and all rows that may correspond to that individual have the same value for Disease. The attacker infered with 100% sure the disease the person has, even though he/she doesn’t know the exact record. [MKGV07]
4. Background knowledge attack - An attacker may have external information about an indi-vidual that removes possible rows for that indiindi-vidual. [MKGV07]
Example: Suppose an attacker knows that an individual corresponds to one of three rows in the anonymized database. From those 3 rows, a column Disease only has two distinct values. So, the attacker infers that the individual has one of two diseases. However, the attacker also knows that the individual does not have one of the diseases. So, it is only left one disease and with this background knowledge, the attacker finds out the disease a specific individual has.
2.2.2 (δ , k) anonymity
(δ , k) anonymity is a simple model that extends k-anonymity, described on Section 2.2.1. In this model, no sensitive value may have a frequency greater than δ within an equivalence class. [Pod11]
2.2.3 kmanonymity
As well as (δ , k) anonymity, kmanonymity is a simple model that extends k-anonymity, described on Section2.2.1. This model intends to ensure that it is required more than m information values about an individual to identify less than k records. [GAZ+14]
Name Glucose Values John 75, 80, 100, 95 Joshua 150, 144 Mary 80, 95, 125 Peter 144, 130, 125 Andrew 75, 130, 125 Table 2.3: Original glucose table
Now suppose an attacker knows that John had the following glucose values: 75mg/dl and 100mg/dl. Even if all names are removed from Table 2.3, the attackers knows exactly which record corresponds to John. To prevent from this to happen, consider the 22-anonymity Table2.4. In this new table, even with those two informations about John, the attacker can not identify which record corresponds to John, having 3 possibilities.
Name Glucose Values
* [70-99], [70-99], [100-130], [70-99] * [130-150], [130-150]
* [70-99], [70-99], [100-130] * [130-150], [130-150], [100-130] * [70-99][130-150], [100-130]
Table 2.4: 22-anonymity glucose table
2.2.4 Bayes-Optimal Privacy
Every attacker has different levels of knowledge that can be useful to discover sensitive informa-tion. Bayes-Optimal Privacy is based on conditional probabilities, which are used to represent the knowledge of an attacker. This model has the concept of prior and posterior belief, which are the beliefs of an attacker knowing a sensitive value. In order to guarantee Bayes-Optimal Privacy the difference between prior and posterior belief must be low. [Ker13,Pod11]
2.2.5 `-diversity
`-diversity was proposed by Machanavajjhala et al. in 2007 and it is based on the Bayes-Optimal Privacy (explained in Section2.2.4).
This principle, defined at Definition2.2.2, tells that there must be a minimum of ` distinct values on sensitive attribute for every group of QIDs. When this is achieved, a table is said `-diverse. [MKGV07]
k-anonymity is satisfied by this privacy model with k = `. This happens because every tuple of quasi-identifiers must contain ` distinct sensitive values and as consequence every tuple appear at least ` times. [BFW+10]
Definition 2.2.2 A table is `-diverse if every equivalence class in that table has a minimum of ` distinct values on sensitive attributes. [MKGV07]
In a `-diverse table, it is needed at least `-1 samples of information about an individual to discover the row that corresponds to that individual. [Pod11]
Age Sex Disease
37 Male Heart Disease 34 Female Cancer
31 Male HIV
24 Female Cancer 26 Male Heart Disease Table 2.5: Patient’s diseases table
Age Sex Disease [30-40] * Heart Disease [30-40] * Cancer
[30-40] * HIV
[20-30) * Cancer [20-30) * Heart Disease Table 2.6: 2-diversity patient’s diseases table
Example 2.2.2 Consider the patients records shown in Table2.5and the corresponding 2-diverse table in Table2.6.
The sensitive attribute is Disease and there are 3 possible values for this attribute - "Heart Disease", "Cancer" and "HIV".
In the 2-diverse version, 2 equivalence classes were created: 1. Age: [30-40] and Sex: *
2. Age: [20-30) and Sex: *
Each of these equivalence classes contain at least 2 distinct sensitive values: 1. First equivalence class contains "Heart Disease", "Cancer" and "HIV" 2. Second equivalence class contains "Heart Disease" and "Cancer" This satisfies`-diversity model with ` = 2.
2.2.5.1 Instantiations
Machanavajjhala et al. proposed three variations of `-Diversity:
1. Distinct `-diversity - this definition is the simplest one and just ensures that at least ` distinct values appear in each equivalence class. [LLV07]
2. Recursive (c-`)-diversity [LLV07] - this variation tries to decrease the difference between the frequency most common values appear and the frequency less common values appear. 3. Entropy `-diversity - this variation takes advantage of the concept of Entropy and it tells
that a table is Entropy `-diverse when each equivalence class has an entropy greater than log(`).
2.2.5.2 Limitations
`-diversity has some weaknesses and limitations that are easily understood. [LLV07]
First of all, `-diversity may be hard to accomplish in some cases. In a dataset with huge amount of records and a sensitive attribute which has a value that rarely appears, `-diversity is really hard to achieve and the information loss would be large. [LLV07]
Second, `-diversity can not prevent attribute disclosure. Even with ` distinct values on each equivalence class for the sensitive attribute, it is not ensured that the values present in the sensitive attribute are not similar or related. If they are similar, then the attacker may infer a valid disclosure. For example, imagine that all sensitive values for an equivalence class are related to a heart disease. Although all values are distinct, the attacker can infer with 100% sure that an individual has heart problems.
2.2.6 t-closeness
t-closeness, proposed by Ninghui Li et al. in 2007 [LLV07], can be considered as an improvement of `-diversity.
This model intends to reduce the level of detail of sensitive attributes. To achieve this, the distribution of a sensitive atrribute within an equivalence class and within the entire table must not have a distance of more than t.[LLV07] To calculate this distance between the two distributions, it is used the earth mover’s distance (EMD). [RTG00,arxd]
As in the previous models, for a table to be closeness, every classes must accomplish t-closeness.
This privacy model is difficult to achieve in some situations and it is even not needed to protect data in most cases as other simpler privacy models are enough.
2.2.7 δ -Presence
δ -Presence evaluates the probability of an individual to be in the generalized table. A dataset is δ -present when this probability is within the range (δmin, δmax). [NC10]
This model is intended to prevent membership disclosure.
2.3
Data models
Data models are another key part when choosing an algorithm for data anonymization. In what concerns anonymization, data can be of two main types - Numeric, Categorical. Recently, ap-peared the concept of Set-valued as data model.
2.3.1 Numeric data
An attribute is considered numeric data for anonymization purposes when it there is strong order among elements. An example of numeric data is Income (Tables2.7and2.8).
Person Id Income 1 30K 2 35K 3 27K 4 23K 5 38K
Table 2.7: Income table
Person Id Income 1 [30K - 40K] 2 [30K - 40K] 3 [20K - 30K] 4 [20K - 30K] 5 [30K - 40K]
Table 2.8: Anonymized income table
2.3.2 Categorical data
An attribute is considered categorical data for anonymization purposes when there is no reasonable order among elements. An example of categorical data is Disease. This type of data is often arranged hierarchically and can be described at different levels of generality. An example of generalization of this type of data can be seen at Figure2.1.
Figure 2.1: Categorical data generalization
2.3.3 Set-valued data
Set-valued data tells that an individual is associated to a set of items. A logical record has the form (indId, {item1, . . ., itemm}) and the representation is conceptually easy: Pindividual= item1, item2,
item3. [HN09,TMK08]
2.4
Approaches to anonymity
In order to achieve anonymity on a database, there are three main approaches: 1. Generalization
2. Suppression 3. Perturbation
2.4.1 Generalization
Generalization is one of the approaches to anonymize data. This approach consists on replacing values with more general values. These generalized values are a set of values that contain the initial value. [KT12] In other words, there is a hierarchy of values where each value has a corresponding value in the next generalization level. When a generalization is done, a value is replaced by its correspondent in the next most general level. [TMK11]
Generalizations tend to have less information loss when there are more levels in their hierarchy. This is easily understandable as more levels mean that each level has greater precision.
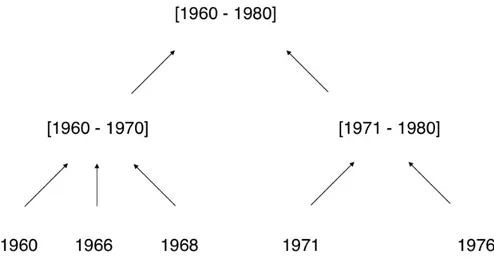
In Figure2.2 there is an example of a generalization hierarchy where the most generalized level is [1960-1980] and the less generalized is a singular year.
Figure 2.2: Date generalization
Generalization can be done using two distinct methods: global recoding and local recod-ing. Global recoding maintains more consistency along the dataset, while local recoding enables anonymization with smaller loss of information.
Local recoding generalizes individual tuples. In this model, each column is generalized inde-pendently to its hierarchicaly generalization. For example, if the weight 65kg is present in several records of the database, it may be generalized to [60-65], to another value or even not suffer changes. [KT12,XWP+06]
Global recoding generalizes every individual to a unique value of the hierarchicaly gener-alization. In other words, each value in the dataset is generalized the same way. [KT12] For example, if the weight 65kg appears in several records, it will be generalized the same way in all records.
These two models for generalization are represented in Table2.10and2.11. Let’s focus on the value 65 for attribute weight. We notice that:
1. in Table2.10it was generalized into [60-65] (row with Id = 1) and into [65-70] (row with Id = 2). This is possible because it was used local recoding model.
2. in Table2.11it was only generalized into [60-70]. This happens because it was used global recoding model and, so, all values in the table were generalized the same way.
Id Weight 1 65 2 65 3 60 4 57 5 69 6 55
Table 2.9: Original table
Id Weight 1 [60-65] 2 [65-70] 3 [60-65] 4 [55-60] 5 [65-70] 6 [55-60]
Table 2.10: 2-anonymization table with local recoding Id Weight 1 [60-70] 2 [60-70] 3 [60-70] 4 [55-60] 5 [60-70] 6 [55-60]
Table 2.11: 2-anonymization table with global recoding
2.4.2 Suppression
Suppression consists in entirely removing a value from the dataset. This can be seen as a general-ization, where the value is generalized to suppressed. This occurs when each value generalization increments until no more generalization can occur and the value needs to be suppressed to not include it in the database. [Swe02a]
In Figure 2.3 there is an example of an hierarchy where the last level of generalization is suppression. Suppression happens when generalization is not sufficient to achieve k-anonymity.
2.4.3 Perturbation
Data perturbation is a technique in which data in a record is changed to an insignificant value. This approach makes harder to re-identify any individual as data is perturbated and changed. Also, even if an individual record is re-identified, there is no certainty that the data on that record corresponds to the initial data.
Micro-aggregation is a method that allows perturbation. This method consists in grouping values in small aggregations and replacing those values with the average of the small aggregation the value is in. The records that belong to the same aggregation will be represented with the same value in the released dataset. [ASMN12]
Id Weight 1 90 2 87 3 85 4 68 5 66 6 69 7 50 8 50 9 52
Table 2.12: Weight table
Example 2.4.1 The above table presents the weight of 9 people. Using the method micro-aggregation for data perturbation, 3 groups are created: [1-3], [4-6] and [7-9]. With these 3 groups created, the average for the weight is calculated for each group: [87.33], [67.67] and [50.67]. Then, the perturbated table looks like the following:
Id Weight 1 87.33 2 87.33 3 87.33 4 67.67 5 67.67 6 67.67 7 50.67 8 50.67 9 50.67
Table 2.13: Perturbed weight table
There are several other techniques for perturbation, which depend on many factors - level of anonymity required, type of data, ... -, such as: [ASMN12]
1. Data swapping - consists in altering records by swapping values with selected pairs of records (swap pairs),
2. Adding noise - consists in adding a random value to all values in the attribute to be protected. 3. Post-Randomization Method (PRAM) - for protecting categorical variables. This method swaps the values for selected variables based on a transition matrix, which specifies the probabilities to swap a value with another value. An example of this technique is better explained in Example2.4.2. [GKWW98,TMK13]
Example 2.4.2 PRAM
Suppose the variable disease has 4 possible values: "cancer", "hiv", "diabetes" and "heart disease". A possible transition matrix could be
Ai, j= 0.2 0.6 0.1 0.1 0.8 0.2 0 0 0.1 0.5 0.2 0.2 0 0.3 0.5 0.2
The transition matrix has the probabilities of each variable being swapped by another. From the matrix it is possible to notice that pcancer,diabetes= 0.1; phiv,diabetes= 0, which means that "hiv"
will not be swapped by "diabetes" in any situation.
2.5
Quality Metrics
As explained in the previous section, anonymization is reached by generalizing and suppressing data. However, with this generalization and suppression, datasets tend to lose quality and infor-mation. It is important not to forget about the quality of generated data and try to find a balance between anonymization and information loss.
Quality metrics of anonymization are an important part of this process and can be expressed in the form of information loss and utility. [RK13] This section discusses and shows up some of these quality metrics.
2.5.1 Discernibility metric (DM)
Discernibility was proposed by Bayardo et al in 2005 [BA05] and penalizes for suppressed rows. [KPK15]
The discernibility metric penalizes each record based on how many records are indistinguish-able from it in the anonymized dataset. The dataset total penalization is equal to the sum of the penalization of each record. The penalty applied to a tuple is:
Penalty(r) =
j, Unsuppressed tuple is present in an equivalence class with size j |D|, Suppressed tuple. |D| is dataset’s size
Penalties on this metric are applied based on the equivalence classes size and not based on in-formation loss. It supposes that the inin-formation loss will be proportional to the size of equivalence classes. [Pod11]
2.5.2 Classification metric (CM)
Each record is assigned a class label as an additional attribute. It evaluates and tries to ensure that generalization and anonymization does not weaken too much the distinction of existing classes using QID.
Classification metric, according to Iyengar, was proposed as: [Byu07,BKBL07]
CM=
∑
allrows Penalty(r) N (2.2) Where: r= specific record N = number of records.Penalty(r)may assume one of the following values:[Byu07,BKBL07]
Penalty(r) =
1, r6= majority class in the equivalence group 1, ris suppressed
0, other situations
(2.3)
This metric is suitable when the anonymized data is used to train a classifier. [GKKM07]
2.5.3 Precision (Prec)
The Precision (Prec) metric was introduced by Sweeney [Swe02a] with the aim to evaluate the information loss. This information loss is evaluated taking into account the amount of modification of a table caused by anonymization.
This metric consists in calculating the ratio between number of generalizations made and total of possible generalizations. This ratio gives the information loss for a specific variable. [EEDI+09, Swe02a]
2.5.4 Normalized Certainty Penalty (NCP)
This metric, which was proposed by Jian Xu et al. [XWP+06], is based on the approximation of generalized entries to the original ones, which allows to evaluate the information loss caused by anonymization process.
As it is based on the approximation of entries to the initial values, it separates the definition into the two main types of data models: numeric and categorical. Each of these main types has a specific way to be evaluated.
2.6
Algorithms
Anonymization is a topic of intense investigation and a wide range of algorithms to achieve anonymization have already been proposed. These algorithms use privacy models, explained at Section2.2, to check whether a data set is anonymized or not. The most recent algorithms also take into account the quality of generated data.In order to find the best balance between informa-tion loss and anonymizainforma-tion, they use quality metrics to evaluate the quality of data and select the best solution according to that metric.
In this section, some algorithms used to achieve k-anonymity will be stated.
2.6.1 DataFly
Datafly was proposed by Latanya Sweeney and aims to provide anonymity in clinical data [Swe98].
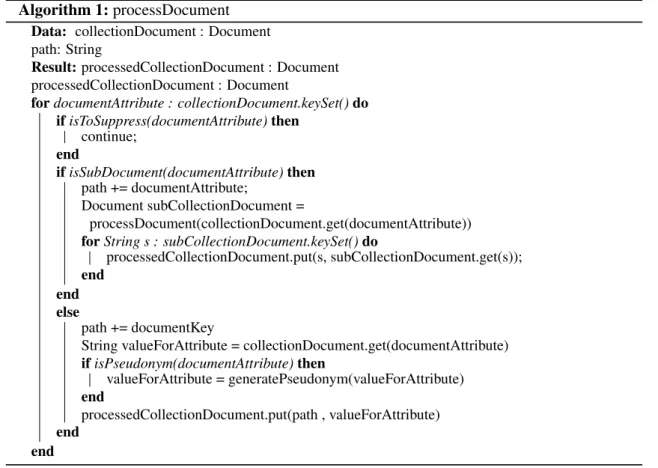
Figure 2.4: Datafly pseudocode (Source: [Swe02a])
This algorithm achieves k-anonymity using global recoding as generalization model and it is based on greedy algorithms. It has few steps, which are demonstrated in Figure2.4[Swe02a]:
1. Construct list of frequencies for QID, where each element corresponds to at least one tuple on the database.
2. While there are elements that occur k or more times in the list, the attribute with more distinct values in that list is generalized.
3. After generalization loop ends, any element on the list that occurs less than k times is sup-pressed.
Datafly has good performance when compared to other algorithms and solutions. However, this algorithm performs unnecessary generalizations and as so, it does not provide an optimal solution even the solution satisfies k-anonymity. This is one of the biggest problems with Datafly as it may generate a solution with high information loss. [Swe02a,EEDI+09,ARMCM14]
2.6.2 Optimal Lattice Anonymization (OLA)
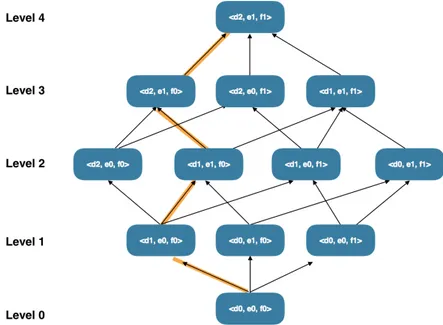
Generalization hierarchies for the QIDs tuples can be represented as a lattice, in which each node represents a possible version of the database The optimal solution for the anonymization prob-lem is within the lattice, corresponding to one of the nodes. Figure 2.5 represents a lattice of generalizations for QID <d, e, f>.
As more generalization is done and as we go up on lattice levels, the suppression percentage reduces. Although it seems confusing at first glance, it is easy to understand. When we are at level 0 of the lattice, there is no generalization. In this case, the quantity of distinct values is enormous and a lot of suppression is needed in order to achieve k-anonymity. However, at level 3 of a lattice some generalization has already been made. In this new case, there is not as much distinct values as in the first scenario and it is easier to achieve k-anonymity without much suppression.
Another thing that is important to understand is that suppression only affects a single record, while generalization affects many records at the same time. For this reason, suppression is pre-ferred to generalization. However, suppressing all records increases information loss and, due to this situation, it is important to impose a limit on suppression. This limit will be referenced as MaxSup. [EEDI+09]
A node in the lattice will be globally optimal if the amount of suppression is less than MaxSup, if it fulfills k-anonymity requirements and if it has the best value for the selected quality metric.
OLA algorithm, proposed by El Emam et al. [EEDI+09], aims to find the optimal solution for the anonymization problem, which corresponds to a node in the lattice - the node with minimal information loss. This algorithm consists in three steps, described in the article published by its author [EEDI+09]:
1. Find all nodes that achieve k-anonymity on each generalization strategy. This is done by using binary search method.
2. The node with lower level in the lattice and that achieves anonymity is selected as k-minimal node in the generalization strategy. The worst case occurs when the only node that achieves k-anonymity corresponds to suppression (higher level). From example in Figure 2.5, suppose nodes <d2, e1, f0> and <d1, e1, f0>, which are in the same generalization strategy, are k-anonymous. As <d1, e1, f0> is below <d2, e1, f0> in the lattice, it means that <d1, e1, f0> will have less information loss. In this generalization strategy, k-minimal node is <d1, e1, f0>.
Figure 2.5: Lattice of generalizations and generalization strategy (orange trace).
3. After having all k-minimal nodes, they are compared with each other based on the quality metric selected. This allows to compare the information loss of each node. The optimal solution is the node with less information loss.
In order to minimize computational costs and improve performance, the algorithm imple-ments predictive tagging. This predictive tagging consists in tagging whether or not a node is k-anonymous after being analysed. Suming to this, in a generalization strategy, all nodes above a k-anonymous node are marked as k-anonymous in the lattice. This tagging allows to not evaluate several times the same node and as consequence, improve algorithm’s performance. [EEDI+09]
2.6.3 Incognito
Incognito algorithm was developed and proposed by LeFevre et al. in 2005. [LDR05] This anonymization algorithm also uses the concept of lattice to find the optimal solution, exposed in Section2.6.2.
Incognito creates lattices for all possible combinations of QIDs, starting from single-attributes subset and stopping with lattices of all QID. [LDR05] I.e., suppose we have 3 QIDs - a, b and c. Incognito would start by creating a lattice for each a, b and c. After analysing these lattices, 3 new ones would be created and analysed: (a, b), (a,c) and (b,c). And so on until no more combinations are possible. At the end, the k-minimal nodes are compared and the best one is returned.
Similarly to OLA algorithm, Incognito uses tags to prevent nodes from being evaluated several times. When a node is k-anonymous in a generalization strategy, it is tagged as so and all nodes with higher height in the lattice are also tagged as k-anonymous.
Another optimization this algorithm takes advantage of occurs when a node does not achieve k-anonymity on smaller subsets. If a node does not achieve k-anonymity on smaller subsets, then
Figure 2.6: Incognito pseudocode (Source: [LDR05]).
for larger subsets that node will also not achieve k-anonymity. This means that some nodes in lattices of larger subsets of QID can be suppressed and so, the computational effort for evaluating it is less than if all nodes were needed to be evaluated. [EEDI+09,LDR05]
For evaluating the lattice, Incognito algorithm starts from the bottom nodes and moves up-wards using Breadth-First Search. Similarly to OLA, as it passes on the nodes, it tags them to prevent repeated evaluations.
The pseudo-code, taken from the article published by its author [LDR05], for this algorithm can be seen in Figure2.6and the evolution of the algorithm on the lattice can be seen on Figure 2.7, taken from [KPE+12].
Figure 2.8: Flash algorithm lattice example (Source: [KPE+12]).
2.6.4 Flash
Flash algorithm was proposed by Kohlmayer et al.. This anonymization algorithm uses the same concept of lattice as Incognito (Section2.6.3) and OLA (Section2.6.2). According to its author, this algorithm goes over the lattice using a bottom-up breadth-first approach. [KPE+12]
This algorithm uses a greedy depth-first strategy and the lattice is traversed vertically. It uses predictive tagging to reduce the number of nodes to be examined. Flash algorithm iterates through every node and finds the path from that node to the next node that only has tagged successors. If that condition is not fulfilled, the path will be from the node in question to the top node. The cre-ated path is checked using binary search so that anonymous nodes are tagged and non anonymous are added to the heap. After the path check is done, the algorithm continues to next iteration using the heap nodes.
The algorithm ends when top level is the starting node. The algorithm pseudo-code can be found in Figures2.9,2.10and2.11, retrieved from its original article. [KPE+12]
Figure 2.10: CheckPath(Path, Heap) (Source: [KPE+12]
Figure 2.11: FindPath(Node) (Source: [KPE+12]
2.7
Tools
Currently, there are already some tools to solve the problem of anonymization. In this section, some tools will be addressed:
• PARAT [Inc] is the leading de-identification software. However, it is closed source and the available information to the public is limited.
• Open Anonymizer [ope] is an open source tool which uses k-anonymity to protect sensitive data by generalizing data records.
• µ-Argus [uar] is a non-commercial software that implements many techniques. However, this tool is closed-source.
• UTD Anonymization Toolbox [utd] is a toolbox with several anonymization methods imple-mented.
• ARX [arxd] is an open source graphical user interface (GUI) software for data anonymiza-tion.
From the list of tools presented above, UTD Anonymization Toolbox and ARX will be covered in more detail nextly, as they are free and open source and may be important for future work on this project.
2.7.1 UTD Anonymization Toolbox
UTD Anonymization Toolboxis a compilation of several anonymization methods, implemented by UT Dallas Data Security and Privacy Lab. As it is public, this toolbox can be used and extended by researchers for their own purposes. [KIK16]
Currently, it supports 3 privacy models by implementing 6 different anonymization methods: [utd]
• Datafly • Incognito
• Incognito with `-diversity • Incognito with t-closeness • Anonymity
• Mondrian Multidimensional k-anonymity 2.7.1.1 Input, Output and Configuration
This toolbox not only has the implementation of the anonymization methods, but also allows to anonymize some dataset if desired.
In order to anonymize data, the input needs to follow some rules and some configuration may be needed.
As input, the toolbox only supports unstructured text files, such as CSV files. It is needed to specify the input file organization in the configuration file.
The output and input format are, by default, the same.
The configuration is done using a XML structured file and starts with a root node config. The root node has at most 5 children, which have all the information required to handle the provided input: [KIK16]
1. input 2. output
3. List of identifiers (represented as id) - optional parameter 4. List of QIDs (represented as qid)
5. List of sensitive attributes (represented as sens) - optional parameter
An example of a configuration file, taken from this tool manual, can be seen in Figure2.12.
2.7.1.2 Extending
This toolbox can be extended in order to add new methods. Every method extends the abstract class Anonymizer. To extend this toolbox and implement new anonymization methods, it is simply required to extend Anonymizer class and represent data according to the documentation. [KIK16] This is useful for researchers as they can add their own methods and implementations to the toolbox, using the already existing structure.
2.7.2 ARX - Powerful Data Anonymization Tool
ARX is an open source software for data anonymization. With an intuitive cross-platform GUI, as seen in Figure 2.13, this tool can handle and anonymize large datasets. It also has a public
Figure 2.12: Configuration file, taken from UTD Anonymization Toolbox manual (Source: [KIK16])
API, which may be useful for other developers when implementing other software that require anonymization.
Figure 2.13: ARX main frame
This tools has several features and supports: [arxa] • Risk-based anonymization.
• Well known privacy models and quality metrics
• Data transformation using generalization, suppression and microaggregation. • Data utility analysis.
2.7.2.1 Anonymization Process
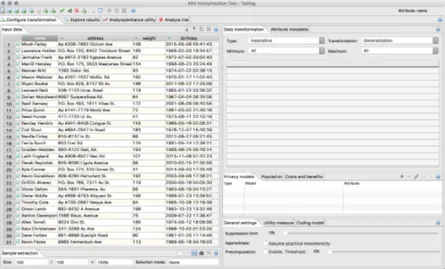
The anonymization process centers in finding a balance between privacy and data utility for re-search. With this concern, ARX tool is based on a 3-step data anonymization process, represented in Figure2.14: (1) configuring the model, (2) exploring the results and (3) comparing and ana-lyzing the output and input data. Each step of this process will be explained in more detail in the following sections.
Figure 2.14: ARX workflow (Source: [arxc]
2.7.2.2 Importing Data and Configuring
Currently, ARX supports importing three types of data: (1) character-separated values files (also known as CSV), (2) Microsof Excel spreadsheets and (3) relational database management systems (RDBMSs).
According to the documentation provided, before anonymizing the imported data, it is impor-tant and necessary to configure at least: [PKLK14,arxa]
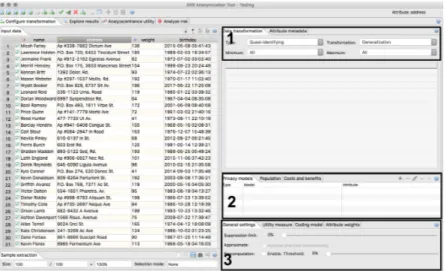
1. generalization hierarchies - hierarchies for each attribute should be created manually. The tool has a feature to help the creation of those hierarchies, which is represented in Figure 2.15.
2. attribute properties - assign a type to each attribute. The type can be quasi-identifier, iden-tifier, sensitive and insensitive.
3. privacy criteria - select the privacy model to use from the list of supported models. 4. utility metrics - select the metric to measure data utility from the list of available metrics. 5. suppression limit - maximum number of records that can be suppressed. It is recommended
Figure 2.15: ARX hierarchy wizard
Attribute properties assignment, privacy criteria, utility metrics and suppression limit selection are represented in Figure2.16.
Figure 2.16: (1) Attribute properties configuration. (2) Privacy models selection. (3) Utility met-rics and suppression limit.
2.7.2.3 Solution Exploration
After a solution space has been classified, ARX tool allows the user to browse the lattice created for the anonymization problem by using the exploration perspective, which is shown in Figure 2.17. This is useful to understand which and why a node was selected and applied by the tool in the anonymization process.
Figure 2.17: ARX solution exploring
2.7.2.4 Transformed data analysis
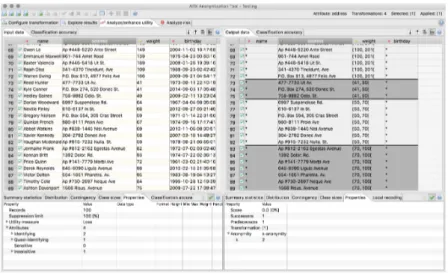
Although ARX automatically finds the optimal solution according to the selected metric for data utility, it provides a useful feature to analyse anonymized data and find out the utility of the trans-formed data. [PKLK14]
This feature, which is represented in Figure 2.18, allows to compare statistical properties, frequency distributions of values and many other metrics between input and output datasets.
Figure 2.18: ARX anonymization result analysis
2.7.2.5 Access via API
Another important part of this tool is the public API it provides. This API aims to provide anonymization methods to other software system and facilitate the implementation of software that needs anonymization.
In Figure2.19, taken from the documentation [arxb], a Unified Model Language (UML) class diagram for the API can be seen, with seven packages:
1. Data utility metrics - access available quality metrics
3. Utility analysis - calculate utility of anonymization 4. Risk analysis - evaluate the risk of anonymization 5. Privacy criteria - access available privacy models
6. Data import & specification - import data from available input data types 7. Hierarchy creation - access to methods to create hierarchies.
These packages are accessed through the core classes ARXConfiguration, ARXAnonymizer and ARXResult. Through all these packages that integrate the API, it is possible to use all features provided by the GUI.
Figure 2.19: ARX tool API uml (Source: [arxb])
2.7.2.6 Limitations
Although it is a complete tool, with several privacy models and analysis features implemented, it has some limitations that must be referenced.
Firstly, as input data it only supports CSV files, Microsoft Excel and RDBMS. NoSQL databases are currently getting more and more common in the world of technology. Consequently, this is a notorious limitation of this tool as it does not provide support for NoSQL databases, such as MongoDB.
Secondly, according to Gkoulalas-Divanis [GDL15], as it uses globally-optimal search strat-egy, it can only handle small search spaces. This limitation, however, is relative as it also depends on several factors, such as hierarchies size.
In third place, it currently does not implement methods to support set-valued data, such as km-anonymity. This is not, however, a big problem as most of anonymization problems can be solved using the available models.
In fourth and last place, although it is possible to anonymize clinical data with this tool, it requires high level of configuration to achieve good results. This may be an obstacle as it re-quires good knowledge about clinical data and even with this knowledge, it may be complicated to achieve the best and desired results as the configurations and restrictions may not be easy to implement using the available GUI.
2.8
Clinical Data Anonymization
The HIPAA (Health Insurance Portability and Accountability Act) Privacy Rule is a set of rules needed to be taken into account when anonymization of clinical datasets is done. Two basic methods for datasets anonymization are defined by this rule. The first requires the suppression of a set of attributes. The second balances the quality of data and anonymization, using methods such as k-anonymity. [PKLK14]
In order to anonymize clinical data and based on the requirements of the HIPPA Privacy Rule, TransCelerate BioPharma Inc. proposed some models to handle specific types of data which will be shown in the following section. [Inc13,oHS+03]
2.8.1 Recoding Identifiers
Individuals have an identifier associated to them in the database, which may allow to identify the individual in case of knowledge of this identifier in the database. A possible approach to anonymize this type of data is by recoding it. This can be done by generating random identifiers which are not possible to reverse.
When recoding identifiers, it is important to use the same new identifiers across all datasets in order to maintain relationship in the database. [Inc13]
2.8.2 Names, Contact information and Identifiers
According to the HIPAA Privacy Rule, all attributes that directly link a record to an individual should be removed or set to blank. [oHS+03]
2.8.3 Age and Date of Birth
According to HIPAA Privacy Rule, individual’s date of birth and ages above 89 may compromise anonymity. This way, date of birth should be removed and ages above 89 should be aggregated into a single category - "above 89". [oHS+03]
2.8.4 Other Dates
Dates not related to age can also compromise anonymity. In order to guarantee anonymization, these dates should be generalized or be replaced with a new date. This new date is calculated using an offset. [Inc13] For example, suppose the date 1Jan2017 was in the database. To anonymize this data using an offset of 90 days, a possible new date could be 18Fev2017.
2.8.5 Medical dictionaries
Medical dictionaries correspond to specific clinical data, such as names of diseases and names of drugs. Although it does not directly identify an individual, it may lead to a correct identification of an individual when associated to external data.
In order to reduce the risk of identity disclosure, this type data must be generalized during the process of anonymization.
2.9
Summary
In this chapter, several concepts as well as related work were introduced within the context of encryption and anonymization of data in general and clinical data in more specific. This helped to clearly understand the topic of this dissertation.
We conclude that there is a large amount of research on this topic and that there are already many privacy models and algorithms to achieve anonymity. Some scientific project and third-party tools have already been implemented to help to solve this privacy problem. These tools allow to anonymize relational databases, missing on the support to MongoDB databases, which are being increasingly used.
Regarding clinical data, there are some data hierarchies, restrictions and data types (see Sec-tion2.8) that must be configured and taken into account when performing anonymization. Some restrictions on data, which must be enforced when sharing data, were already defined by HIPAA. From this, we can conclude that, in order to anonymize clinical data with the already existing tools, it is required high level of configuration in order to implement these restrictions and add all needed hierarchies.
Solution
On Chapter 1 it is possible to conclude that sharing clinical data for research purposes brings several benefits to pharmaceutical industry and clinical research in general. However, data sharing brings several and critical problems that must be solved in order to allow the continuing of clinical data share. Among these problems, the most important one, and in which all problems are based on, is privacy disclosure.
In previous chapter, the state of the art on anonymization of data was analyzed. Given this, it is possible to conclude that many models and algorithms already exist in the fields of data anonymization. Some tools also have already been developed. However, some of these tools are not free and the others are not focused on clinical data and require a lot of configuration, which is not easy. Another point that is a big obstacle is the type of database those tools support - none of them support MongoDB, which is in huge expansion.
With this in mind, in this chapter, a tool for anonymizing clinical data will be presented, with the objective of providing the user with an easy to use interface that allows to quickly and effectively anonymize clinical data that is stored in a MongoDB database. This tool will take into account specific clinical data hierarchies and restrictions (e.g. diseases and drugs).
3.1
Requirements
As specified on Section 1.2, this dissertation aims to create a solution that makes clinical data sharing possible. From a database, the solution must create a new anonymized database.
The solution must be applied to a Mongo database and should be scalable enough to be inde-pendent of the collection structure. All documents inside the MongoDB collection must have the same well defined structured in order to be anonymized. The results must be stored into a new MongoDB collection with the same structure as the non-anonymized collection.
Another important requirement is the amount of data the solution can handle. The system in which the process will run has limited amount of memory and processing units. So, the amount of data that can be stored into memory during the process is limited. The solution must be able to handle the maximum amount of data.
To reduce the amount of data, the anonymization process will run in a monthly basis. This is a way of clustering the process without losing too much information. However, in order to find out correlations between data, it must be possible to associate data inside the database. This means that there must be a way to tell that two different records correspond to the same individual, but without compromising its privacy.
In terms of speed and time of operation, it is essential to maintain a balance between the amount of resources required by the system and the processing time. This means that the solution should have good response time but this response time must be balanced with the resources needed to complete the process.
In order to handle different collections, the solution must be easily configured by the database administrator. This configuration must contain information about the attributes used by the anonymiza-tion process, such as QID and sensitive attributes.
3.1.1 Database
As already specified before, the anonymization process will be applied to a MongoDB database and more in specific to two different collections: Prescribed Medications and Consumptions.
Documents inside a collection must have a fixed structure over that entire collection. 3.1.1.1 Indexes
As specified in Section3.1, the anonymization process will run with a monthly periodicity. This means that the query to fetch the collection to anonymize will only use the creation date field. For this reason, it is recommended to add an index to the attribute DateReg. This attribute contains the date the record was created at. Both collections have this attribute.
3.2
Architecture
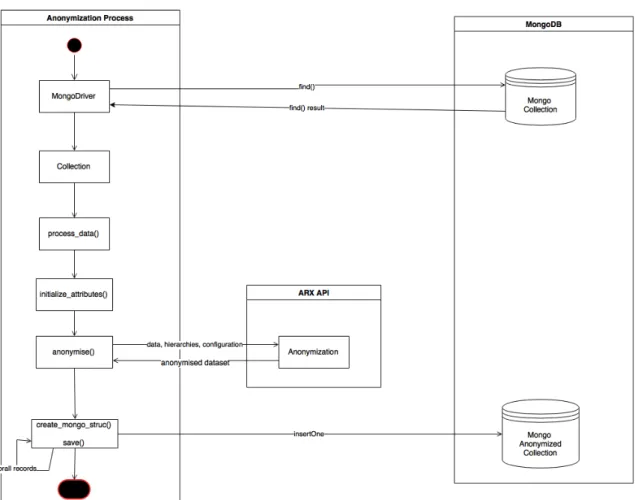
From the requirements analysis, three main modules were defined for a better final solution: 1. Anonymization process itself - receives as parameter a configuration file, a MongoDB
collection and the destination collection. This module fetches the collection, anonymizes it and stores the anonymized version in a new mongo collection. Returns a result string with information about the process (quality metrics, elapsed time, ...).
2. Anonymization GUI app - a simple GUI that allows to: (1) connect to a MongoDB col-lection, (2) create a configuration file with an easy to use GUI, (3) start the anonymization process, (4) preview the anonymized version and (5) export results to a MongoDB collec-tion.
3. Anonymization web service - allows the administrator to automate the anonymization and view some useful analytics. By using this module, the anonymization will run automatically every month or week and the results can be later analysed using the web dashboard.
![Figure 2.4: Datafly pseudocode (Source: [Swe02a] )](https://thumb-eu.123doks.com/thumbv2/123dok_br/19203228.954700/41.892.249.688.572.800/figure-datafly-pseudocode-source-swe-a.webp)
![Figure 2.19: ARX tool API uml (Source: [arxb])](https://thumb-eu.123doks.com/thumbv2/123dok_br/19203228.954700/52.892.104.748.440.877/figure-arx-tool-api-uml-source-arxb.webp)