w w w . r b h h . o r g
Revista
Brasileira
de
Hematologia
e
Hemoterapia
Brazilian
Journal
of
Hematology
and
Hemotherapy
Original
article
Validation
of
interphase
fluorescence
in
situ
hybridization
(iFISH)
for
multiple
myeloma
using
CD138
positive
cells
Renata
Kiyomi
Kishimoto
a,∗,
Sarah
Lee
Vaughan
Vulcani
de
Freitas
a,
Cristina
Alonso
Ratis
a,
Daniela
Borri
a,
Roberta
Sitnik
a,
Elvira
Deolinda
Rodrigues
Pereira
Velloso
a,baHospitalIsraelitaAlbertEinstein,SãoPaulo,SP,Brazil bUniversidadedeSãoPaulo(USP),SãoPaulo,SP,Brazil
a
r
t
i
c
l
e
i
n
f
o
Articlehistory:
Received10November2015 Accepted26January2016 Availableonline23February2016
Keywords:
Cytogenetics Multiplemyeloma Plasmacells
Fluorescenceinsituhybridization CD138cells
a
b
s
t
r
a
c
t
Background:Multiplemyelomaisaplasmacellneoplasmwithacquiredgenetic abnormali-tiesofclinicalandprognosticimportance.Multiplemyelomadiffersfromotherhematologic malignanciesduetoahighfractionoflowproliferatingmalignantplasmacellsandthe paucityofplasmacellsinbonemarrowaspirationsamples,makingcytogeneticanalysis achallenge.Anabnormalkaryotypeisfoundinonlyone-thirdofpatientswithmultiple myelomaandinterphasefluorescenceinsituhybridizationisthemostusefultestfor study-ingthechromosomalabnormalitiespresentinalmost90%ofcases.However,itisnecessary tostudythegeneticabnormalitiesinplasmacellsaftertheiridentificationorselectionby morphology,immunophenotypingorsorting.Otherchallengesaretheselectionofthemost informativeFISHpanelanddeterminingcut-offlevelsforFISHprobes.Thisstudyreports thevalidationofinterphasefluorescenceinsituhybridizationusingCD138positivecells, accordingtoproposedguidelinespublishedbytheEuropeanMyelomaNetwork(EMN)in 2012.
Method:Bonemarrowsamplesfrompatientswithmultiplemyelomawereusedto stan-dardizeapaneloffiveprobes[1qamplification,13q14deletion,17pdeletion,t(4;14),and t(14;16)]inCD138+cellspurifiedbymagneticcellsorting.
Results:Thistestwasvalidatedwithalowturnaroundtimeandgoodreproducibility.Fiveof sixsamplesshowedgeneticabnormalities.Monosomy/deletion13plust(4;14)werefound intwocases.
Conclusion: Thistechniquetogetherwithmagneticcellsortingiseffectiveandcanbeused intheroutinelaboratorypractice.Inaddition,magneticcellsortingprovidesapureplasma cellpopulationthatallowsothermolecularandgenomicstudies.
©2016Associac¸ ˜aoBrasileiradeHematologia,HemoterapiaeTerapiaCelular.Published byElsevierEditoraLtda.Allrightsreserved.
∗ Correspondingauthorat:HospitalIsraelitaAlbertEinstein,LaboratóriodeTécnicasEspeciais–Citogenética,Av.AlbertEinstein,627/701, 05651-901SãoPaulo,SP,Brazil.
E-mailaddress:renata.kishimoto@einstein.br(R.K.Kishimoto). http://dx.doi.org/10.1016/j.bjhh.2016.01.005
Introduction
Multiplemyeloma(MM)isadiseasecharacterizedbyclonal proliferation ofplasma cells (PCs) in bone marrow, which leadstobonemarrowfailure,skeletallesions,suppressionof normalimmunoglobulinsynthesisandproductionofa mono-clonalprotein.1Thisdiseaseaccountsforbetween10%and 15%ofhematologicalcancers.Themedianageatdiagnosisis 60years,andtheevolutionisheterogeneous,withthesurvival timevaryingfromafewmonthstomorethanadecade.2,3
MM shows acquired genetic abnormalities of clinical importance. In about half of the cases, the initial genetic process involves a reciprocal translocation between the immunoglobulinheavy (IgH) gene(14q32)and many target genesincludingCCND1(11q13),FGFR3/MMSET(4p16)andMAF
(16q23).4,5
Thestudyofcytogeneticabnormalitiesbykaryotypingis limited becauseof the lowmitotic index ofthe malignant PCs.6–8 Only 20–50% of the cases show clonal abnormal-ities by G banding karyotype. However, the presence of hypodiploidyormonosomyofchromosome13predictspoor survival.9–13
Molecular studies show that most MM cases present genetic abnormalities with interphase fluorescence in situ
hybridization(iFISH) beingthemostuseful cytogenetictool fortheirinvestigation.7,14,15 However,iFISHtestingrequires previous identification or selection of PCs by morphology, immunophenotypingorsorting.Cellselectionusingthe anti-CD138antibodycanbeperformedusingmagneticcolumns orsorting.Themajorlimitationofthisapproachisthe con-siderable loss of cells during the purification process. In the cytoplasmic immunoglobulin (Clg) fluorescence in situ
hybridization (FISH) technique, PC detection is carried out using fluorescentanti-Kappa or anti-Lambda antibodies in thePCcytoplasmandanalysisisperformedonlyusingthis population.4,5,16
Theother challengesinMMtestingwithFISHareprobe selection, the determination of cut-off levels and number
ofPCstobescored.TheEuropeanMyelomaNetwork(EMN) hasorganizedtwoworkshopsoniFISHinMM.In2012,they published some technical recommendations herein tran-scribedfromthepaper:
(1) Materialshouldbepartofthefirstdrawoftheaspirate; (2) Samplesshouldbesentatsuitabletimestoallowforthe
lengthyprocessingprocedure;
(3) Most importantly, PCs must bepurified or specifically identified;
(4) Cut-offlevelsshouldberelativelyconservative:10%for fusion or breakapart probes, and 20% for numerical abnormalities;
(5) Informativeprobesshouldbecombinedforbesteffect; (6) Inspecialistlaboratories,asingleexperiencedanalystis
consideredadequate;
(7) Atleast100PCsshouldbescored;
(8) Essentialabnormalitiestotestforaret(4;14),t(14;16)and 17p13deletions;
(9) Suitablecommercialprobesshouldbeavailablefor clini-callyrelevantabnormalities;
(10) Theclinicalreportshouldbeexpressedclearlyandmust statethepercentageofPCinvolvedandthemethodused foridentification”.4
Objective
ThisstudyaimedtostandardizeaniFISHpaneltestforMMfor itsincorporationinlaboratoriesasaroutinecytogenetictest.
Methods
ThisresearchwasevaluatedbytheEthicsCommitteeofthe Hospital(SGPPnumber169913–“Validationoflaboratorytests forClinicalPathologyLaboratory”).Thestudywasconducted inaccordancewiththeHelsinkiDeclarationasrevisedin2008.
Anti-CD138 + Anti-dextran Antibody complex
Magnetic beads
with dextran Magnetic column
Sample Add lysis solution Add cocktail Add magnetic beads Insert tube in magnetic column
Wash
Figure1–Magneticcellsorting(MACS)ofCD138+cellsusingthekitEasySepTMHumanWBandBMCD138PositiveSelection
50
50
100
150
200
250
50
100
1
50
FSC-A
CD138 PE-A CD138 FITC-A
(x 1000) (x 1000)
CD45 P
erCP-Cy5-5-A
200
2
50
10
2
10
3
10
4
1
0
5
10
2
10
3
10
4
1
0
5
10
2
10
3
10
4
1
0
5
10
2
10
3
10
4
1
0
5
100 150 200 250
(x 1000) (x 1000) (x 1000)
CD45
(x 1000) (x 1000)
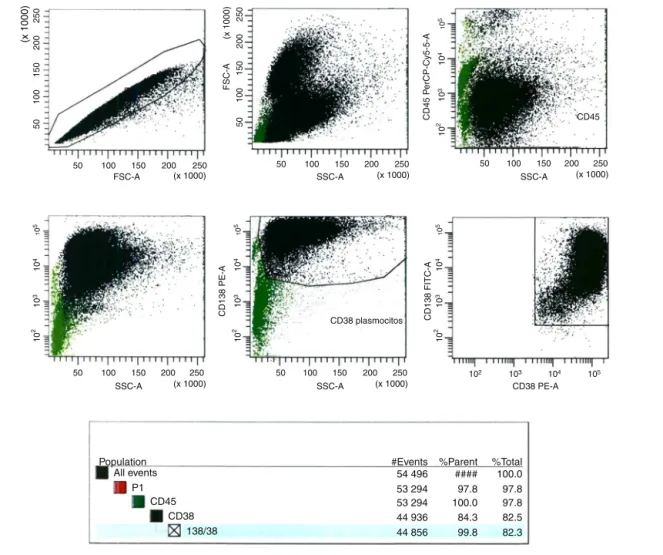
Population All events
P1 CD45
CD38 138/38
#Events %Parent %Total 54 496 #### 100.0 53 294 97.8 97.8 53 294 100.0 97.8 44 936 84.3 82.5 44 856 99.8 82.3
50 100 150 200 250 50 100 150 200 250
50 100 150
SSC-A SSC-A
CD38 plasmocitos SSC-A
FSC-A SSC-A
CD38 PE-A
200 250 50 100 150 200 250 102 103 104 105
Figure2–FlowcytometryanalysisofMACSenrichment.
Twelve bone marrow samples from patients diagnosed with MM received at the Cytogenetic Laboratory of a pri-vate Hospital in Sao Paulo from March to October 2014 wereselectedforthisvalidation.Theprocessisdescribedin Figures1–5.
MagneticcellsortingofCD138+cells
Thisfirst stepwasperformed using the“EasySepTM Human
WB”, “BM CD138 Positive Selection Cocktail” and “EasySepTM
WholeBloodMagneticParticles”(StemcellTechnologiesTM)kits. Theprocess started lessthan 12hafterthesamplewas collected,atroomtemperaturefollowingthemanufacturers’ protocol.Attheend oftheprocess,the tube wasremoved fromthemagneticcolumnandtheselectedmaterialwas re-suspendedin200Lofwashedsolution,whichwasusedafter thisstepforflowcytometryanalysisandtheharvest proce-dure(Figure1).
Flowcytometryanalysisofmagneticcellsorting
enrichment
Atleast100cells(countedinaNeubauerchamber)ofthe mag-neticcellsorting(MACS)enrichmentsamplewereevaluated
Hypotonic solution Fixative
Centrifuge and remove the supernatant
Incubation at 37ºC
FISH
(Fluorescent
in situ
hybridization)
One drop of fixed cells in slide
Add probes labeled with fluorochromes Target DNA
Probe and target DNAs are denatured using a high temperature incubation
Probes anneals to
target DNA Nucleus is stained
blue with DAPI
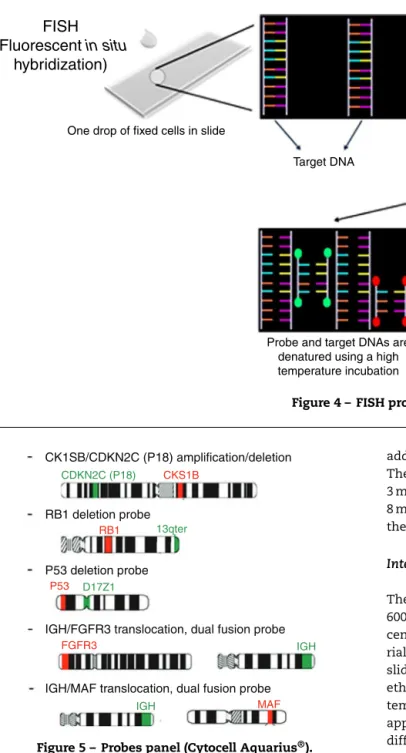
Figure4–FISHprocedure.
CK1SB/CDKN2C (P18) amplification/deletion
RB1 deletion probe
P53 deletion probe
IGH/FGFR3 translocation, dual fusion probe
IGH/MAF translocation, dual fusion probe
CDKN2C (P18)
13qter
D17Z1
IGH
IGH
CKS1B
RB1
P53
FGFR3
MAF
Figure5–Probespanel(CytocellAquarius®).
byflowcytometry.Anti-CD138-FTIC(B-A38clone,Exbio), anti-CD38-PE(T16clone,Immunotech)andanti-CD45-PE-Cy5(J33 clone,Immunotech)monoclonalantibodies(MoAb)wereused toidentifybonemarrowPCs.Flowanalysiswascarriedoutin FACSCantoIIequipment (Becton,DickinsonandCompany). Data analysiswasperformedusing theBDFACS Diva soft-ware(Becton, Dickinsonand Company, version 6.1.3, USA) (Figure2).
Harvestprocedure
Threemillilitersofhypotonicpotassiumchloridesolution(KCl –0.075mol/L)wereadded tothe MACSenrichmentsample andincubatedfor16minat37◦C.Afterthistime,1mLof fixa-tivesolution(Carnoy’ssolution:3:1methanol/aceticacid)was
added andthe tubewas centrifugedat1500rpmfor8min. Thesupernatantwasdiscarded;materialwasre-suspendedin 3mLofCarnoy’ssolutionandcentrifugedagainat1500rpmfor 8min.Thelatterprocedurewasrepeatedtwomoretimesand theresultingpelletwasusedintheiFISHprocedure(Figure3).
Interphasefluorescenceinsituhybridizationprocedure
The pellet obtained in the last step was re-suspended in 600–1000LofCarnoy’ssolutionandcentrifugedinaCytospin centrifuge at 1000rpm for 5min (100–200L of the mate-rialforeachslide).Accordingtothemanufactures’protocol, slideswerepretreatedinthefollowingsolutions:2×SSC,70%
ethanol, 85% ethanol and 100% ethanol for2minat room temperature.TenmicrolitersofCytocellAquarius®probewas appliedonaslideandcoverslipped.Thiswasdonewitheach differentprobe.Co-denaturationoftheprobeandtargetDNA wasperformedonahotplateat75◦Cfor5min.Slideswere incubatedfor12–16hinahumidchamberforhybridization andthenwerewashedin0.4×SSCsolutionat42◦Cfor2min
followedbyasecondwash(2×SSC+0.1%NP40)atroom
tem-peraturefor1min.Nucleiwerecounterstainedwith10Lof DAPI II (Cytocell Aquarius®) and coverslipped. Slides were storedat4◦Cfor10min(Figure4).
Probepanelselectionandscoring
Fiveprobes(CytocellAquarius®)wereselected:Amplification 1qdualcolor;Deletion13q(RB1)dualcolor;Deletion17p13.1 (P53)dualcolor;t(4;14)IGH/FGFR3dualcolor,dualfusionand t(14;16) IGH/MAF dual color, dual fusion. iFISH analysis of probehybridizationwasperformedwitha100×objective
12 Samples
2 Heparin
Ethanol
- Poor quality hybridization
- Good results - Low amount of cells
- Low cell purity
6 Heparin
Hypotonic and fixative
solutions 4 EDTA
Figure6–Studydesign.
twotechnicians(50interphasenucleieach),totaling100cells. Incaseofdiscrepantresults,theanalysisofanother50cells wascarriedoutbyathirdtechnician(Figure5).
Determinationofthecut-offpoints
TheEMNcut-offlevelrecommendationwasfollowed:10%for fusionandbreakapartprobesand20%fornumerical aberra-tions.
Results
Ofthetwelvebonemarrowsamplesusedinthisvalidation test,fourwerecollectedinEDTAandeightinheparinsodium. BonemarrowsamplescollectedinEDTAresultedinlowpurity afterCD138+ selectionandtwosamplesinheparinsodium fixedinethanolresultedinbadhybridization.Therefore,good resultswereobtainedforonlysixsamplescollectedin hep-arinsodiumtreatedwithhypotonicsolution(KCl)andfixedin Carnoy’ssolution.TheseresultsareshowninFigure6.
ThenumberofPCsdetectedbymorphologyonbone mar-rowaspirateslides(not theimpurespecimen)rangedfrom
1.2%to82.8%.Themediantimefromthesamplearrivingin thelaboratorytothestartoftheprocesswas3h(range:2–5h). AfterCD138+ selection,thesamplepurityrangedfrom70to 91%.Athirdanalysiswasperformedfor40%oftheprobes, mainlytoconfirmatypicalsignalsseenfortheIgHprobe.
Karyotype and iFISHresults, described according to the standardsoftheInternationalSystemforHumanCytogenetic Nomenclature(ISCN)2013,17arelistedinTable1.
Fiveofthesixcasesanalyzedshowedabnormalfindingsin iFISH(Figure7).Onlyonecase(Sample11)hadabnormalities foundbybothkaryotyping(Figure8)andiFISH.Both t(4;14) andmonosomy13weredetectedintwocases.Extracopiesof genesandabnormalitiesonchromosome1werealso identi-fied.
Discussion
Anumberofdifferentgenomicabnormalitiesareassociated withMM.However,theirdetectionbykaryotypingandFISH canbelimitedduetothelowproliferativerateofthePCsand thepercentageofclonalcellsinthespecimen,respectively. Abnormalkaryotypes arefoundin20–50%ofpatientswith MM; this rate canbe evenlower whenMM is analyzedat diagnosis.6,7 Karyotypescanshownumericalandstructural aberrations with the most frequent numerical abnormal-ities described being hyperdiploidy in 61–68% of patients, pseudodiploidy in 9–20% and hypodiploidy, monosomy 13
and trisomy or tetrasomy of chromosome 9 in 10–30%.
Concerning non-random structural aberrations, 14q32
translocationsandaberrationsofchromosome1arefoundin 30%and40–50%ofcaseswithabnormalkaryotypes, respec-tively. The results of cytogenetic studies can be improved using longer culture periods (72h)and adding stimulating agentssuchas12-O-tetradecanoylphorbol-13-acetate(TPAor phorbol12-myristate13-acetate)andcytokines(interleukin6 andgranulocyte-macrophagecolony-stimulatingfactor).7
TheselectionofPCsforiFISHanalysisimprovedthe detec-tion of genetic aberrations in MM. Different methods are
Table1–Interphasefluorescenceinsituhybridizationandkaryotyperesultsfromsixsamples.
Sample %ofplasmacellsin bonemarrow
aspirate(%)
%CD138+cellsafter selection(%)
Karyotype FISH
7 40.8 72.6 46,XX20 Trisomy(totalorpartial)of
chromosomes1,4,11,13,17
8 22.0 70.4 46,XX20 del(1p),del(16q)(MAF)
9 Notdone 90.9 46,XX20 Monosomy13,t(4;14)
10 17.6 89.6 46,XY20 1qamplification,
monosomy13,t(4;14)
11 4.80 70
44,X,-Y,add(1)(p34),add(5)(q11.2),del(6)(q23),-7, t(8;11)(q24;q13),-12,add(13)(p11.2),-14,-16,-18,
+der(19)t(1;19)(q23;p13)x2,add(20)(p13), +2mar[2]/46,XY[18]
ExtracopiesofCDKN2C,
CKS1B,P53andFGFR3
A
B
C
E
1q P53 13q
14;16 4;14
D
Figure7–AbnormalitiesfoundinfiveanalyzedsamplesusingtheiFISHtechniqueformultiplemyeloma.(A)1q
amplification(severalredsignals);(B)trisomy17(3redand3greensignals);(C)deletion/monosomy13q(onlyoneredand onegreensignalinonenucleus);(D)4redand4greensignals,suggestingextracopiesofchromosomes4and14;(E) t(14:16):1red,1greenand2fusionsignals.
Figure8–Abnormalmetaphase(Sample11)showing severalabnormalities
(44,X,-Y,add(1)(p34),add(5)(q11.2),del(6)(q23),-7, t(8;11)(q24;q13),-12,add(13)(p11.2),-14,-16,-18,
+der(19)t(1;19)(q23;p13)x2,add(20)(p13),+2mar[2]/46,XY[18]).
describedfortargetingthePCsforiFISHanalysis.Oneis per-formedinunpurifiedspecimens:the analysisiscarriedout usingonlylargemononuclearcells.Thismethodresultsin lowsensitivity,andinvolvesalongtimetotrainanalyststo recognizethePCs,aprolongedanalysistime,acertaindegree ofsubjectivityandlackofreproducibility.5Thesecondisthe CIg-FISHtechnique.Thisanalysisisperformedonmonotypic PCs recognized byimmunofluorescence-labeled light chain antibodies.15Thistechnique,whichalsorequiresalongtime fortraininganddoesnothavegoodreproducibility,wasused intwoBrazilianstudies.18,19Thethirdapproachisbyselecting cellseitherbyflowcytometryorimmunomagneticbead-based PCsorting.4
Thecurrentstudyfollowedthethirdapproachusingthe magneticcellsorting(MACS)techniquegivingtheimpression
thatthismethodispossibleasaroutinelaboratorytestifsome precautionsaretaken.Themagneticcellsortingmustbe per-formedwithinafewhours.Hartmannetal.5reportedatime dependenceforPCenrichment:thepercentageofPCsinthe enrichedpopulationdecreasedsignificantlywiththeageof thespecimen.Thisphenomenonisduetotherapidlossof theCD138markeronPCsafterthecellsaretakenoutofthe bonemarrow20;slidesarestableandcanbestoredforalonger timebeforetheiFISHprocedure.
ThepanelprobeshereinselectedwerebasedontheEMN consensus.1qamplification,t(4;14),t(14;16)and17p13 dele-tion are associatedwithbad prognoses,4,8,12,21 and the13q deletionisstillcontroversial.18Inparticular,t(4;14)(p16;q32) atdiagnosishasbeenshownasabadprognosticmarker,18 eveninsmolderingMM22,23andinpatientswithsymptomatic MMtreatedwithconventionalchemotherapy,whichmaybe amelioratedwithbortezomib-basedcombinations.2
Another important issue is the cut-off levels for FISH probes. Some authors use statistical analysis (B-inv func-tion,averageandstandarddeviation)ofnormalcontrolcases todeterminethecut-offlevels. However,toselectPCsfrom normalbonemarrowisverydifficult.Thisstudyusedthe con-sensuscut-offlevelsdefinedbytheiFISHmyelomaworkshop: 10% for fusion or breakapartprobes and 20% for numeri-calabnormalities.4Nevertheless,ingeneral,badprognosisis associatedwithahigherpositivitysuchas30%fordel(13q) and50%fordel(17q).
AlthoughtheENMguidelinessuggestthatasingle experi-encedanalystisconsideredenoughinspecialistlaboratories, thisstudywasperformedbytwotechnicians(50cellseach).A goodreproducibilitywasobtained,butinsomedoubtfulcases, anextraanalysisbyathirdanalystwasnecessary.
ishigherthanourhistoricaldetectionrateof34% abnormali-tiesfrom2007to2014,whensamplesweresenttoareference laboratorythatperformedFISHanalysisforMMwithoutPC selection(datanotshown,personalcommunication).
SelectionofPCsbyMACScanalsobeanimportanttool forstudyingthegeneticprofileofMMwithnovel methods suchasmicroarray-basedcomparativegenomichybridization (array-CGH), multiplexligation dependent probe amplifica-tion(MLPA)andmassivelyparallelsequencing,althoughthe clinicalrelevance ofthesemethodsstillneedsto bebetter established.24–26
Conclusion
Insummary,theiFISHtechniqueusingPCsortingwithMACS iseffective,hasagoodturnaroundtimeandgood reproducibil-ity.Thetestwasvalidatedandestablishedinthelaboratory routine.Inaddition,thiskindofcellsortingprovidesapure PCpopulationthatsuitableforothermolecularandgenomic studies.
Conflicts
of
interest
Theauthorsdeclarenoconflictsofinterest.
Acknowledgments
WethankNydiaBacal,MDandthebiologistsofthe labora-toryofCytometryandCytogeneticsofHospitalIsraelitaAlbert Einsteinfortheirexcellenttechnicalassistance.
r
e
f
e
r
e
n
c
e
s
1. KyleRA,RajkumarSV.Multiplemyeloma.Blood. 2008;111(6):2962–72.
2. FonsecaR,BergsagelPL,DrachJ,ShaughnessyJ,GutierrezN, StewartAK,etal.InternationalMyelomaWorkingGroup molecularclassificationofmultiplemyeloma:spotlight review.Leukemia.2009;23(12):2210–21.
3. UKMyelomaForum.BritishCommitteeforStandardsin Haematology.Diagnosisandmanagementofmultiple myeloma.BrJHaematol.2001;115(3):522–40.
4. RossFM,Avet-LoiseauH,AmeyeG,GutiérrezNC,LiebischP, O’ConnorS,etal.ReportfromtheEuropeanMyeloma NetworkoninterphaseFISHinmultiplemyelomaandrelated disorders.Haematologica.2012;97(8):1272–7.
5. HartmannL,BiggerstaffJS,ChapmanDB,ScottJM,Johnson KR,GhirardelliKM,etal.Detectionofgenomicabnormalities inmultiplemyeloma:theapplicationofFISHanalysisin combinationwithvariousplasmacellenrichment techniques.AmJClinPathol.2011;136(5):712–20. 6. DewaldGW,KyleRA,HicksGA,GreippPR.Theclinical
significanceofcytogeneticstudiesin100patientswith multiplemyeloma,plasmacellleukemia,oramyloidosis. Blood.1985;66(2):380–90.
7.ZandeckiM,LaïJL,FaconT.Multiplemyeloma:almostall patientsarecytogeneticallyabnormal.BrJHaematol. 1996;94(2):217–27.
8.FonsecaR,BarlogieB,BatailleR,BastardC,BergsagelPL, ChesiM,etal.Geneticsandcytogeneticsofmultiple myeloma:aworkshopreport.CancerRes.2004;64(4): 1546–58.
9.TricotG,BarlogieB,JagannathS,BracyD,MattoxS,Vesole DH,etal.Poorprognosisinmultiplemyelomaisassociated onlywithpartialorcompletedeletionsofchromosome13or abnormalitiesinvolving11qandnotwithotherkaryotype abnormalities.Blood.1995;86(11):4250–6.
10.SmadjaNV,BastardC,BrigaudeauC,LerouxD,FruchartC, GroupeFranc¸aisdeCytogénétiqueHématologique. Hypodiploidyisamajorprognosticfactorinmultiple myeloma.Blood.2001;98(7):2229–38.
11.Debes-MarunCS,DewaldGW,BryantS,PickenE, Santana-DávilaR,González-PazN,etal.Chromosome abnormalitiesclusteringanditsimplicationsfor pathogenesisandprognosisinmyeloma.Leukemia. 2003;17(2):427–36.
12.StewartAK,BergsagelPL,GreippPR,DispenzieriA,GertzMA, HaymanSR,etal.Apracticalguidetodefininghigh-risk myelomaforclinicaltrials,patientcounselingandchoiceof therapy.Leukemia.2007;21(3):529–34.
13.KyleRA,RajkumarSV.Criteriafordiagnosis,staging,risk stratificationandresponseassessmentofmultiplemyeloma. Leukemia.2009;23(1):3–9.
14.Pérez-SimónJA,García-SanzR,TaberneroMD,AlmeidaJ, GonzálezM,Fernández-CalvoJ,etal.Prognosticvalueof numericalchromosomeaberrationsinmultiplemyeloma:a FISHanalysisof15differentchromosomes.Blood.
1998;91(9):3366–71.
15.AhmannGJ,JalalSM,JuneauAL,ChristensenER,HansonCA, DewaldGW,etal.Anovelthree-color,clone-specific fluorescenceinsituhybridizationprocedureformonoclonal gammopathies.CancerGenetCytogenet.1998;101(1): 7–11.
16.PutN,LemmensH,WlodarskaI,KoningsP,MoreauY, HagemeijerA,etal.Interphasefluorescenceinsitu hybridizationonselectedplasmacellsissuperiorinthe detectionofcytogeneticaberrationsinplasmacelldyscrasia. GenesChromosomesCancer.2010;49(11):991–7.
17.ShafferLG,McGowan-JordanJ,SchmidM,editors.An internationalsystemforhumancytogeneticnomenclature (2013).Basel:S.Karger;2013.
18.LinardiCC,MartinezG,VellosoED,LealAM,KumedaCA, BuccheriV,etal.Evaluationofchromosomalabnormalitiesby cIg-FISHandassociationwithproliferativeandapoptotic indexesinmultiplemyeloma.BrazJMedBiolRes. 2012;45(11):1074–9.
19.SeggesPA,BraggioE,MaiolinoA,RenaultIZ.Estudoda prevalênciadealterac¸õescitogenéticasempacientescom mielomamúltiplo(MM)porclg-FISH.RevBrasHematol Hemoter.2008;30Supl4:189.
20.JourdanM,FerlinM,LegouffeE,HorvathovaM,LiautardJ, RossiJF,etal.Themyelomacellantigensyndecan-1islostby apoptoticmyelomacells.BrJHaematol.1998;100(4):
637–46.
21.HungriaVT,CrusoeEQ,QueroAA,SampaioM,MaiolinoA, BernardoWM.Guidelinesonthediagnosisandmanagement ofmultiplemyelomatreatment:Associac¸ãoBrasileirade HematologiaeHemoterapiaeTerapiaCelularProject guidelines:Associac¸ãoMédicaBrasileira–2012.RevBras HematolHemoter.2013;35(3):201–17.
abnormalitiesandriskofprogressioninsmolderingmultiple myeloma.Leukemia.2013;27(8):1738–44.
23.RajkumarSV,DimopoulosMA,PalumboA,BladeJ,MerliniG, MateosMV,etal.InternationalMyelomaWorkingGroup updatedcriteriaforthediagnosisofmultiplemyeloma. LancetOncol.2014;15(12):e538–48.
24.WalkerBA,BoyleEM,WardellCP,MurisonA,BegumDB,Dahir NM,etal.Mutationalspectrum,copynumberchanges,and outcome:resultsofasequencingstudyofpatientswith newlydiagnosedmyeloma.JClinOncol.2015;33(33): 3911–20.
25.BoyleEM,ProszekPZ,KaiserMF,BegumD,DahirN,SavolaS, etal.Amoleculardiagnosticapproachabletodetectthe recurrentgeneticprognosticfactorstypicalofpresenting myeloma.GenesChromosomesCancer.2015;54(2): 91–8.