w w w . r b h h . o r g
Revista
Brasileira
de
Hematologia
e
Hemoterapia
Brazilian
Journal
of
Hematology
and
Hemotherapy
Original
article
Identification
of
the
MYST3-CREBBP
fusion
gene
in
infants
with
acute
myeloid
leukemia
and
hemophagocytosis
Francianne
Gomes
Andrade
a,
Elda
Pereira
Noronha
a,
Rosania
Maria
Baseggio
b,
Teresa
Cristina
Cardoso
Fonseca
c,
Bruno
Marcelo
Rocha
Freire
d,
Isis
M.
Quezado
Magalhaes
e,
Ilana
R.
Zalcberg
a,
Maria
S.
Pombo-de-Oliveira
a,∗ aInstitutoNacionaldeCâncer(INCA),RiodeJaneiro,RJ,BrazilbHospitalRegionaldoMatoGrossodoSulRosaPedrossian(HRMS),CampoGrande,MS,Brazil
cHospitalManoelNovais,SantaCasadeMisericórdiadeItabuna(HMN-SCMI),Itabuna,BA,Brazil
dHospitalSantaIzabel,Salvador,BA,Brazil
eHospitaldaCrianc¸aJosedeAlencar(HCB),Brasília,DF,Brazil
a
r
t
i
c
l
e
i
n
f
o
Articlehistory: Received25June2015 Accepted16June2016 Availableonline26July2016
Keywords: Infantleukemia Acutemyeloidleukemia t(8;16)(p11;p13)
MYST3-CREBBP(or,MOZ-CBP) Hemophagocytosis
a
b
s
t
r
a
c
t
Background:AcutemyeloidleukemiapresentingtheMYST3-CREBBPfusiongeneisarare subgroupassociatedwithhemophagocytosisinearlyinfancyandmonocyticdifferentiation. Theaimofthisstudywastodefinetherelevantmolecularcytogeneticcharacteristicsofa uniqueseriesofearlyinfancyacutemyeloidleukemiacases(≤24monthsold),basedonthe presenceofhemophagocytosisbyblastcellsatdiagnosis.
Methods:Aseriesof266infantcasesofacutemyeloidleukemiawasthereferencecohortfor thepresentanalysis.Acutemyeloidleukemiacaseswithhemophagocytosisbyblastcells werereviewedtoinvestigatethepresenceoftheMYST3-CREBBPfusiongenebyfluorescence insituhybridization(FISH)andreversetranscriptionpolymerasechainreaction.
Results:Elevencaseswithhemophagocytosiswereidentifiedwithhemophagocytic lympho-histiocytosisbeingruledout.Sixcaseswereclassifiedasmyelomonocyticleukemia,threeas AML-M7andtwoasAML-M2.Infivecases,thepresenceoftheMYST3-CREBBPfusiongene identifiedbymolecularcytogeneticswasconfirmedbyfluorescenceinsituhybridization. AllpatientsreceivedtreatmentaccordingtotheBerlin–Frankfürt–Münsteracutemyeloid leukemiaprotocolsandonlyoneoutofthefivepatientswiththeMYST3-CREBBPfusion geneisstillalive.
Conclusions: Ourfindings demonstrate thatthepresence of hemophagocytosisin acute myeloid leukemia was not exclusively associated to the MYST3-CREBBP fusion gene. Improvementsinmolecularcytogeneticsmayhelptoelucidatemorecomplexchromosomal rearrangementsininfantswithacutemyeloidleukemiaandhemophagocytosis.
©2016Associac¸ ˜aoBrasileiradeHematologia,HemoterapiaeTerapiaCelular.Published byElsevierEditoraLtda.ThisisanopenaccessarticleundertheCCBY-NC-NDlicense
(http://creativecommons.org/licenses/by-nc-nd/4.0/).
∗ Correspondingauthorat:ProgramadeHematologia-OncologiaPediatrico,InstitutoNacionaldeCâncer(INCA),RuaAndréCavalcanti,
37,20231-050RiodeJaneiro,RJ,Brazil.
E-mailaddress:mpombo@inca.gov.br(M.S.Pombo-de-Oliveira).
http://dx.doi.org/10.1016/j.bjhh.2016.06.005
Introduction
Distinct cytogenetic subgroups of acute myeloid leukemia (AML) have been associated with age-specific frequencies and the incidence of unbalanced aberrations; in particu-lar complex karyotypes increase sharply with age.1 AML presenting the reciprocal translocation (8;16)(p11;p13) that generates the MYST3-CREBBP (former named as MOZ-CBP) fusiongeneismostlyobservedinadultpatients.2Thefusion of the MYST3 and CREBBP genes occurs when both show histone acetyltransferase activities leading to the activa-tionofseveraltargets involvedintranscriptionalregulation and cell cycle control.2–4 The evidence of AML with the MYST3-CREBBPfusiongene inchildrenwasreportedbythe InternationalBerlin–Frankfurt–Munster(I-BFM)studygroup.5 Sixty-twopediatricAMLwereidentifiedinwhichkaryotype recordsrevealed t(8;16)(p11;p13)intheAMLobservedatan earlyage,monocytedifferentiation[French–American–British classification(FAB)AML-M5]andpresenceof hemophagocy-tosis;allofwhichareassociatedwithverypooroutcomes.5 Furthermore,theMYST3-CREBBPfusiongeneassociatedwith disseminated intravascular coagulation and high mortality rates was observed in a series of French AML patients.6 Theseparticularclinical,cytological,cytogenetic,and molec-ular characteristics of AML with MYST3-CREBBP led to the suggestionofaunique categoryinthe World Health Orga-nization (WHO) classification due to the poor prognosis.7 Among the clinical spectrum conditions, hemophagocytic lymphohistiocytosis (HLH) should be included as differen-tialdiagnosis. However,HLH presentsphagocyte activation causedbyimmunedisordersthatcompromiseTcell/natural killercellsandthenormalmonocyte-macrophagelineage.8
Anaccuratecaseidentificationrequirestheevaluationof morphological, cytogenetic and molecular features follow-ingcorrelationofobtainedparameters,includingserological tests.Inthisstudy,theavailabilityofauniqueseriesofearly onset AML cases prompted us to search for AML- MYST3-CREBBPcases and todefine relevant molecularcytogenetic characteristics.
Methods
Subjects
A series of 266 infant AML (i-AML) cases enrolled in the BrazilianCollaborativeStudyGroupofInfantAcuteLeukemia (BCSGIAL)from2003to2012isthereferencecohortandsubject forthepresentanalysis.9Theselectioncriteriawereinfants (≤24monthsold)withadiagnosisofAMLandthepresence ofhemophagocytosisbyleukemicblasts(Figure1). Addition-ally,48i-AMLcaseswithoutthehemophagocyticfeatureinthe diagnosticsampleswererandomlyselectedtocomparewith i-AMLcaseswithhemophagocytosisbyblastcells.
Hemophagocytosiswasdefinedasthepresenceof phago-cytosisofredcells,lymphocytesand/orplateletsonlybyblast cells.Themorphologicalfindings were discussed by physi-cians (RMB, TCCF, BF, IMQM) and cytologists (EPN, MSPO); clinical and laboratorial data were checked in each case fortheconsistencyofinclusioncriteria.Gender, age,white
blood cell count (WBC), hemoglobin levels, platelet count, central nervous system (CNS) involvement, chloroma and cutaneous leukemia,FABclassificationaswellas the pres-enceofhemophagocytosisbyleukemicblastswerecarefully reviewed.ExclusioncriteriaincludedsecondaryAML,down’s syndrome,HLHand/orhemophagocyticsyndromeassociated withimmunedisordersandunexplainedfever.Frozen sam-plesfrombonemarrow(BM)aspirates,peripheralbloodand smearsofi-AMLcaseswereselectedforfurthercytogenetic and molecular studiesaccording totheavailability ofgood biologicalmaterial.
Allchildrenweretreatedoutofclinicaltrials,butfollowing internationalAMLprotocols.
Characterizationofleukemiacells
Leukemia classification ofAMLwas basedon criteria pub-lished by the WHO.7 Thediagnosis of AML-M7 was based on the presenceofCD41/CD61and CD42markerson blast cells identified by immunophenotyping. Karyotypes of BM aspirates were tested before any chemotherapy treatment. Chromosomeswereidentifiedandanalyzedasrecommended bytheInternationalSystemofHumanCytogenetic Nomen-clature(ISCN)2005.10
Reversetranscriptionpolymerasechainreaction
Total RNAfrom BMmononuclearcells atthetime of diag-nosiswaspurifiedusingtheTRIzolreagentaccordingtothe manufacturer’sinstructions(Gibco/BRL,LifeTechnologies,CA, USA).Briefly,2goftotalRNAwasreverse-transcribedusing
theFirst-StrandcDNASynthesisKitTM(AmershamPharmacia BiotechInc.,NJ,USA).TheintegrityofcDNAwasexaminedby amplifyingafragmentoftheGAPDH geneusingpreviously describedprimersandcDNAwasusedastemplatesin sub-sequent polymerase chain reaction (PCR) assays. All cases were investigatedforthe presenceoftheRUNX1-RUNX1T1, CBFˇ-MYH11, BCR-ABL1, MLL-AFF1 and MLL-MLLT3 fusion genes.11–12
DetectionoftheMYST3-CREBBPandreverseCREBBP-MYST3 fusiontranscriptswereconductedasdescribedelsewhere.13 Single PCRreactions todetect MYST3-CREBBPfusion trans-criptstypeI(MYST3exon16-CREBBPexon3)andtypeII(MYST3 exon16-CREBBPexon4),aswellasthetypeICREBBP-MYST3 fusiontranscript(CREBBPexon2-MYST3exon17)were per-formed using the primers listed inTable 1. Asemi-nested reaction,adaptedfromSchmidtetal.,wasrequiredtodetect typeItranscripts.13SamplesfromconfirmedAMLcaseswith MYST3-CREBBP were added as positive controls for type I transcripts.PCRproductsforMYST3-CREBBPtranscripts(type I-II) were separated by electrophoresisin 1.5% agarose gel and subsequently purified using NucleoSpin Gel and PCR Clean-upkits(Macherey-Nagel,VWRInternational,Oslo, Nor-way). Amplicons were mixed with the Big Dye terminator v3.1Kit(AppliedBiosystems)andforwardorreverseprimers and sequencedinan ABI3130xl GeneticAnalyzer (Applied Biosystems).
Figure1–MorphologyofAML-M4withhemophagocytosisbyblastcells.Bonemarrowaspirationstainedby May–Grunwald–Giemsashowsmyeloblastandmonoblastcellswithphagocytosisofredcellsandlymphocytes.
Fluorescenceinsituhybridization
Fluorescencein situ hybridization (FISH)for the MLL rear-rangements was performed at the time of diagnosis with freshbiological materialusing a commercial LSIMLL Dual Color, Break Apart Rearrangement probe (Cytocell Ltd., Cambridge, UK) according to the manufacturer’s instruc-tions.TheMYST3-CREBBPFISHwasperformedininterphase nuclei preparedfrom frozenviable cells of available cases using bacteria-derived artificial chromosome (BAC). These cloneswere retrieved from the human genome high reso-lution BAC re-arrayed clone set available in a web format
(http://bacpac.chori.org) and selected according to physical
andgeneticmappingdatareportedonEnsemblBrowser
web-site(http://www.ensembl.org).DNAwasextractedandprobes
were labeled and hybridized by Blue Genome (Cambridge, UK),withSpectrumOrangeorSpectrumGreenandvalidated as a FISH probe set on normal controls. The clones used wereRP11-231D20(chr8:42184655-42188062)andRP11-108L9 (chr8:41832025-41864392)flanking the MYST3 gene (orange) and RP11-387O21 (chr16:3918191-4104380) and RP11-461A8 (chr16:3663996-3693579) flanking the CREBBP gene (green). Procedureswereperformedaccordingtothemanufacturer’s instructions. The first step was FISH mapping of clones on normal cells from healthy blood donors in order to confirmtheirchromosomallocation.Cut-offvalueswere cal-culatedas6±3%offusiongenesignalsin100–300interphase nuclei.
Ethicalconsiderations
Treatmentwasapprovedbylocallawsandregulationsaswell as bythe InstitutionalReview Boardsof each participating center.Medicalinformedconsentwasobtainedinaccordance withtheDeclarationofHelsinki.Thisstudywasapprovedby theResearchEthicsCommitteeattheInstitutoNacionalde CâncerinRiodeJaneiro,Brazil(CEP/CAEE:186.688).
Results
Theclinical-demographiccharacteristicsoftheelevencases that fulfilled the selection criteria are shown in Table 2. All patientspresentedhepatosplenomegaly andthreewere reportedtohavechloromaandoneCNSdisease.The major-ity of the patients were male (72.7%) with a median age of12 months(range:0–23months). TheWBCcountvaried from 5.7 to111.1×109/Lwith a medianof35.9×109/L; six caseswerediagnosedasmyelomonocyticleukemia(M4/M5), threecasesasAML-M7andtwoasAML-M2.Serologicaltests for viral infections (Epstein–Barr virus,parvovirus B19 and humanimmunodeficiency virus)and coagulation examina-tionswerewithinnormalranges.NoinfectionstriggeringHLH werefoundinanyoftheelevencases.
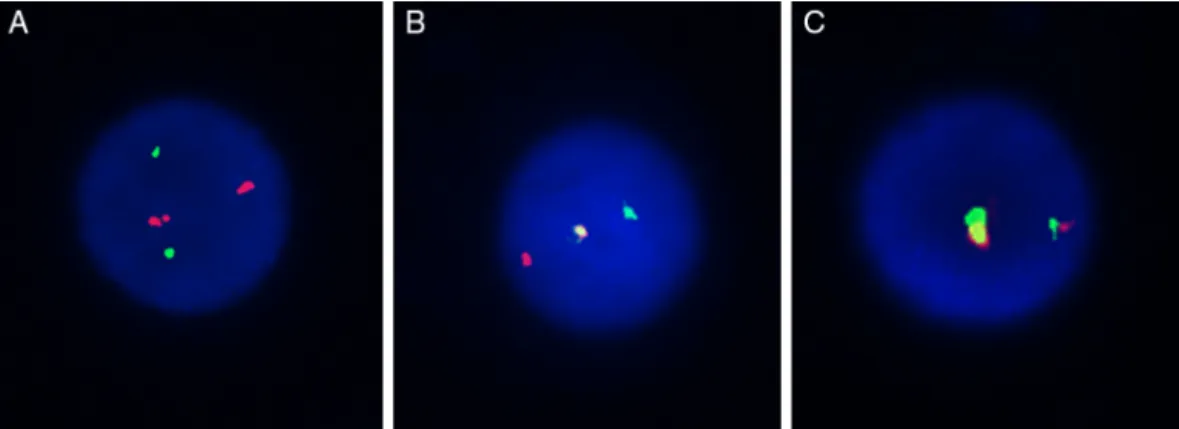
Using the selected BAC clones of the MYST3-CREBBP fusion gene, three types of hybridization patterns were observed (Figure 2). The first, separated signals of the
Table1–Sequencesoftheprimersusedforreversetranscriptionpolymerasechainreaction.
Designation Sequence(from5′to3′) Position
MOZ3558F GAGGCCAATGCCAAGATTAGAAC MOZexon16
CBP1201R GTTGCAATTGCTTGTGTGGGTAC CBPexon5
MOZ3536F CCTTTTGAAGATTCTGACTCCG MOZexon16
CBP404R CCTCGTAGAAGCTCCGACAGTT CBPexon3
CBP96F CGCTCGCTCCTCTCCCTCGCAG CBPexon2
MOZ3953R TGGAAACGATGGGCTCAATGACGC MOZexon17
CBP174F GGGCTGTTTTCGCGAGCAGGTG CBPexon2
Table2–Demographicandclinicalcharacteristicsofselectedi-AMLcases.
Case Age(Mo.) Gender Clinicalfeatures Hb(g/dL) WBC(×109/L) Platcount (×109/L)
Hemoph* FAB Conclusion Outcome
1 12 F Hepatosplenomegaly 5.68 111.1 40.0 + M4 AML-M4
MYST3-CREBPP
Deceased
2 11 M Hepatosplenomegaly 4.0 25.4 26.0 + M5 AML-M5 Alive
3 <1 F Hepatosplenomegaly Chloroma
10.7 8.6 29.0 + M4 AML-M4
MYST3-CREBPP
Deceased
4 12 M Hepatosplenomegaly 4.2 36.8 35.0 + M2 AML-M2
MYST3-CREBPP
Alive
5 18 M Hepatosplenomegaly Chloroma
7.0 5.7 3.0 +/− M5 AML-M5
MYST3-CREBPP
Deceased
6 23 M Hepatosplenomegaly 4.6 42.4 57.0 +/− M7 AML-M7
MYST3-CREBPP
Alive
7 10 M Hepatosplenomegaly Chloroma
10.7 35.0 229.0 + M2 AML-M2 Alive
8 13 M Hepatosplenomegaly 6.2 61.6 45.0 + M5 AML-M5 Alive
9 22 M Hepatosplenomegaly CNSPOS
5.0 25.9 31.0 + M7 AML-M7 Deceased
10 12 M Hepatosplenomegaly 5.5 45.0 100.0 + M5 AML-M5 Deceased
11 11 F Hepatosplenomegaly 7.0 8.5 20.0 + M7 AML-M7 Deceased
F:female;M:male;(*),Hemoph:hemophagocytosis(range:5–27%blastswithphagocytosis);Hb:hemoglobinconcentration;i-AML:infantacute myeloidleukemia;Mo:months;FAB:French–American–Britishclassification;NOS:nototherwisespecified;Plat:plateletcount;WBC:white bloodcellcount.
probe combinations RP11-231D20/RP11-108L9 and
RP11-387O21/RP11-461A8onchromosomes8and 16respectively,
were observed as four different signals: two red and two
greendistinctsignalsconsistentwithnormalchromosomes
(Figure 2A). Second, a single fusion pattern was observed
(onefusion,oneredandonegreen)consideredasarandom co-localizedsignal(Figure2B).Lastly,adualfusionsignalwas foundin15–37%oftheinterphasenucleianalyzedwhichwas consistentwithabreakpointinMYST3andCREBBP(Figure2C). TheRT-PCRtechniquewasperformedin55samples;seven sampleswerefrom i-AMLwithhemophagocytosis(Table3) and48samplesfromi-AMLwithouthemophagocytosis.The RT-PCRpatternwasdifferent toexpected(∼1000bp) inone case,witha∼900bpproductobserved(#1).Infivecases(#3, #5,#6, #7 and #10), RT-PCRwas negative fortype I and II transcripts. Despite accurate mapping ofthe translocation breakpoints,attemptstoamplifythe transcripts,aswell as the CREBPP-MYST3 were not successful. In four cases (#2, #4, #8 and #9), RT-PCR was not performed due to lack of
suitablebiologicalmaterial.Discrepancieswere observedin threecases(#3,#5and#6)inwhichdualfusionsignalswere foundin15–18%and34%oftheinterphasenucleianalyzed and RT-PCR results were negative. In all the i-AML cases withouthemophagocytosis,theRT-PCRresultswerenegative
(Figure3).
As shown in Table 3, the diagnosis of the MYST3-CREBBP fusion gene was based on the FISH results only or combined with the RT-PCR results. Five cases were diagnosedasAML-MYST3-CREBBPandclinicallaboratorial fea-turesare summarized:theypresentedhepatosplenomegaly, skinlesions and/orlocalizedchloroma; hematologicaltests revealed FAB AML-M2, M4, M5, or M7; the presence of hemophagocytosis byblastcells variedfrom5to25%.The immunophenotyping profile showed cells positive for the CD34,CD33/CD13/CD14/CD11b/CD14/CD15,CD64,CD56 anti-gens; in three patients, the blast cells were positive for CD61/CD41a/CD42b/CD56;karyotypingwassuccessfulinfour cases;onecase(#1)revealeda46,XX,der(16),t(16;?)(p13;?)
Figure2–FISHpatternofMYST3-CREBBPprobes.Threetypesofhybridizationpatternswereobserved:tworedandtwo
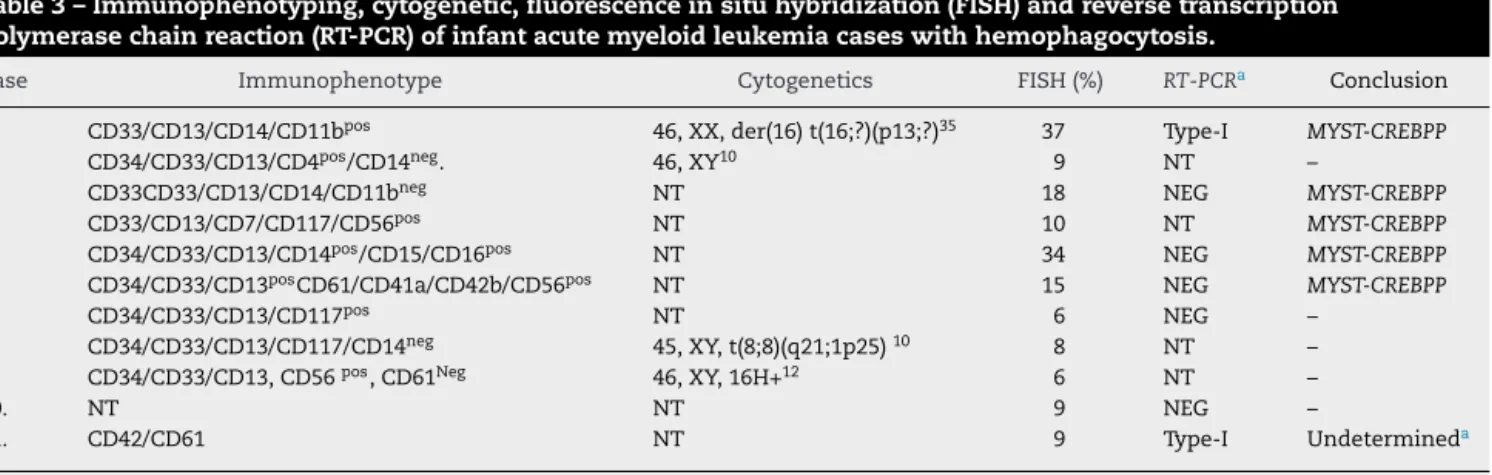
Table3–Immunophenotyping,cytogenetic,fluorescenceinsituhybridization(FISH)andreversetranscription polymerasechainreaction(RT-PCR)ofinfantacutemyeloidleukemiacaseswithhemophagocytosis.
Case Immunophenotype Cytogenetics FISH(%) RT-PCRa Conclusion
1. CD33/CD13/CD14/CD11bpos 46,XX,der(16)t(16;?)(p13;?)35 37 Type-I MYST-CREBPP
2. CD34/CD33/CD13/CD4pos/CD14neg. 46,XY10 9 NT –
3. CD33CD33/CD13/CD14/CD11bneg NT 18 NEG MYST-CREBPP
4. CD33/CD13/CD7/CD117/CD56pos NT 10 NT MYST-CREBPP
5. CD34/CD33/CD13/CD14pos/CD15/CD16pos NT 34 NEG MYST-CREBPP
6. CD34/CD33/CD13posCD61/CD41a/CD42b/CD56pos NT 15 NEG MYST-CREBPP
7. CD34/CD33/CD13/CD117pos NT 6 NEG –
8. CD34/CD33/CD13/CD117/CD14neg 45,XY,t(8;8)(q21;1p25)10 8 NT –
9. CD34/CD33/CD13,CD56pos,CD61Neg 46,XY,16H+12 6 NT –
10. NT NT 9 NEG –
11. CD42/CD61 NT 9 Type-I Undetermineda
–:Considerednegative;NT:nottested;NEG:Negative.
a UndeterminedbecausetheFISHcut-offvalueswerecalculatedas6±3%offusiongenesignals,anddiscordantresultwiththeRT-PCR,the
type-Ifragmentlengthwasnotasexpected.
Figure3–AgarosegelimagesofRT-PCRforMYST3-CREBBPfusiongenes.GelA,showspositivereactionsforthe MYST3-CREBBPfusiongene.Samples2(A)and19(B)arethepositivecontrolsineachreaction.Sample1(AandB)isthe
negativecontrol(H2Oonly).Samples3and5(A)arefrompatients#1and#11.IngelB,allsamplesarenegativeforthe
MYST3-CREBBPfusiongene;M:standardmarker(100basepairs)
withoutidentifiedpartnersontheshortarmofchromosome 16.
NoneoftheelevenAMLcasespresentedwiththeR UNX1-RUNX1T1, CBFb-MYH11, BCR-ABL1, MLL-AFF1, MLL-MLLT1, KRAS,FLT3orc-KITmutations.
ThepatientsreceivedAMLtreatmentaccordingtotheBFM AML-2004protocolandonlyoneoutoffivepatientswiththe AMLMYST3-CREBBPfusiongeneisstillalive.
Discussion
ChromosomalabnormalitiesinchildhoodAMLarefrequent; theMYST3-CREBBPrearrangement,however,isnot.1,17 Here, wereport forthe firsttime the presenceofMYST3-CREBBP rearrangement in five out of eleven (36.4%) AML cases withhemophagocytosis foundin aBrazilian i-AML cohort. Themorphologicalobservationofhemophagocytosiswasan importantvariablefortheselectioncriteriatoinvestigatethe MYST3-CREBBPfusiongene.Clinically,thesecasesappearto havedistinctdiseasemanifestationswithskinnodules,CNS involvementand chloroma.1,5,18 As pointed out by Hatano et al., other chromosomal abnormalities in AML, such as t(16;21)(p11;q22),karyotypes involving the 8p11 breakpoint, t(8;19)(p11;q32),complexrearrangementsandother chromo-somal translocations are associated with the presence of
hemophagocytosis by blast cells.19 This supports our data showinganabsenceoftheMYST3-CREBBPfusiongeneinfour casesinthisstudy.Accordingtotheliterature,inthe major-ity ofpatientsdescribed withmyelomonocytic morphology, thepresenceofCD56cellularexpressionpredictsan associa-tionwithhemophagocytosisandinvolvementoftheleukemia cutis.1,5,19–22
Thetechnique used toidentifythis chromosomal alter-ation was FISH because conventional karyotyping was not alwaysavailable.Failuretoobtainmitosiswasapitfall. Multi-colorkaryotypingtechnologiessuchasmulticolor-FISHwould certainlyelucidatesuchsubtlechromosomalrearrangements ifmitosisweresuccessfullyobtained.Basedonhematological signs,wechosetocarryouttheFISHmethodfollowedby RT-PCRasalaboratorialstrategytosearchfortheMYST3-CREBBP fusiongene.TheFISHanalysisshowedafusionsignalabove thecutoffvalueforthespecificMYST3andCREBBPprobeson interphasenuclei,suggestingthepresenceofaMYST3-CREBBP fusionchimerainfivecases.Thisfindingindicatesapossible generationofMYST3-CREBBP,sincebothchromosomeregions representedbytheclonesthatcontaintheMYST3andCREBBP genesappearedco-localized.Inonecase,RT-PCRforthetype I fusiontranscriptfollowed bydirectsequencingshowed a
adultswithMYST3-CREBBPAML.16,23Lowexpressionor insta-bilityofthechimerictranscripts24andRNAdegradationmight explaintheabsenceofamplificationbyRT-PCR.
Recently, Panagopoulos et al. described an AML with hemophagocytosisandwithtwotranslocationswith break-pointsthatsuggestothercandidategenesdifferenttoMYST3 andCREBBP.25Theystudiedthepatients’leukemiccellsnot only by karyotyping, FISH and RT-PCR, but also using the modern RNA-seqtechniqueand programsthat are specific forfusiongenes.Interestingly,thetechniquesinitiallyfailed to detect the biologically important MYST3-CREBBP fusion, althoughitwasmanuallyretrievablefromtheraw sequenc-ingdata,suggestingthatadditionalinformationaboutclinical, morphological,andmolecularcytogeneticfeaturesshouldbe takenintoaccountwhensearchingfornewlydescribedcrucial fusiongenesintypicalhematologicmalignancies.25
One important point should be discussed is related to AML-MYST3-CREBBP and the differential diagnosis of a hemaphagocytic syndrome such as HLH, which is a severe hyperinflammatory condition with clinical symp-tomsthatincludefever,cytopenias,hepatosplenomegaly,and hemophagocytosis.26However,thishemophagocytosisinBM ismorphologicalinbenignmacrophages.HLH,when occur-ringinyoungchildren,isassociatedwithinheritedgenetic defectsanddiagnostic criteriacombinebothbiological fea-tures, includingnaturalkiller cell activity andhigh-soluble interleukin-2-receptorlevels.27
AMLcasesyoungerthantwoyearsoldwith hemophago-cytosis should be investigated for the presence of the MYST3-CREBBPandotherchromosomalalterations.Inthe I-BFMAMLstudygroup,morethan50%oftheMYST3-CREBBP caseswerefoundininfantsand thefrequencyof congeni-talcaseswassignificantlyhigher.5Oneofourcasesdescribed hereinwascongenitalleukemia.Someauthorsconsiderthat congenital AML-MYST3-CREBBP may be a self-limiting dis-ease reaching spontaneous remission. A ‘watch-and-wait’ policyshouldbeconsideredincongenitalpatientswithmild clinical symptoms provided that close long-term monitor-ing is used.5,28–30 However, casesfrom our cohort suffered fromaggressivediseasewithdismaloutcomes.Interestingly, the genomic landscape of childhood AML-MYST3-CREBBP has a specific signature clustered close to AML with MLL rearrangements.5,31,32Thesimilarityconsistsinthe character-isticpatternofup-regulationofHOXA9,HOXA10,andcofactor MEIS1anddown-regulationofotherhomeoboxfamilygenes.32 Thehigh frequency ofAML-MYST3-CREBBP in infants(≤24 months) and congenital casessupportthe hypothesis that leukemia occurs duringthe in utero lifeand as inthe MLL rearrangementsmodeltheyshouldbeexplored forabetter understandingofAMLleukemogenesis.
Conflict
of
interests
Theauthorsdeclarenoconflictsofinterest.
Acknowledgements
Theauthorsthankthephysicians’partoftheBrazilian Col-laborativeStudyGroupofInfantAcuteLeukemia(BCSGIAL)
fromdifferentBrazilianregionsforsupportingtheprojectby sendinggeneralandclinicaldataofi-AML.
TheauthorsaregratefultoDr.TarsisPaivaVieiraforFISH technicalsupport,BrunodeAlmeidaLopesforimageedition andDr.GerhardFukaforcriticalandEnglishrevisionofthe manuscript.TheauthorsarealsoindebttoDr.OskarHaas fromAnnaKinderspital,MedicalUniversityVienna,Vienna, AustriawhokindlyprovidedtheAML-MYST3-CREBBPsample touseaspositivecontrols.
MSPOwassupportedbyFAPERJ(#E-26/101.562/2010).
r
e
f
e
r
e
n
c
e
s
1.BacherU,KernW,SchnittgerS,HiddemannW,HaferlachT,
SchochC.Population-basedage-specificincidencesof
cytogeneticsubgroupsofacutemyeloidleukemia.
Haematologica.2005;90(11):1502–10.
2.HaferlachT,KohlmannA,KleinHU,RuckertC,DugasM,
WilliamsPM,etal.AMLwithtranslocationt(8;16)(p11;p13)
demonstratesuniquecytomorphological,cytogenetic,
molecularandprognosticfeatures.Leukemia.
2009;23(5):934–43.
3.KatsumotoT,AikawaY,IwamaA,UedaS,IchikawaH,Ochiya
T,etal.MOZisessentialformaintenanceofhematopoietic
stemcells.GenesDev.2006;20(10):1321–30.
4.BorrowJ,StantonVPJr,AndresenJM,BecherR,BehmFG,
ChagantiRS,etal.Thetranslocationt(8;16)(p11;p13)ofacute
myeloidleukemiafusesaputativeacetyltransferasetothe
CREB-bindingprotein.NatGenet.1996;14(1):33–41.
5.CoenenEA,ZwaanCM,ReinhardtD,HarrisonCJ,HaasOA,de
HaasV,etal.Pediatricacutemyeloidleukemiawith
t(8;16)(p11;p13),adistinctclinicalandbiologicalentity:a
collaborativestudybythe
International-Berlin-Frankfurt-MunsterAML-studygroup.
Blood.2013;122(15):2704–13.
6.GervaisC,MuratiA,HeliasC,StruskiS,EischenA,LippertE,
etal.Acutemyeloidleukemiawith8p11(MYST3)
rearrangement:anintegratedcytologic,cytogeneticand
molecularstudybythegroupefrancophonedecytogénétique
hématologique.Leukemia.2008;22(8):1567–75.
7.VardimanJW,ThieleJ,ArberDA,BrunningRD,BorowitzMJ,
PorwitA,etal.The2008revisionoftheWorldHealth
Organization(WHO)classificationofmyeloidneoplasmsand
acuteleukemia:rationaleandimportantchanges.Blood.
2009;114(5):937–51.
8.DelavigneK,BérardE,BertoliS,CorreJ,DuchayneE,DemurC,
etal.Hemophagocyticsyndromeinpatientswithacute
myeloidleukemiaundergoingintensivechemotherapy.
Haematologica.2014;99(3):474–80.
9.Pombo-de-OliveiraMS,KoifmanS,VasconcelosGM,
EmerencianoM,deOliveiraNovaesC.Developmentand
perspectiveofcurrentBrazilianstudiesontheepidemiology
ofchildhoodleukemia.BloodCellsMolDis.2009;42(2):121–5.
10.ShafferLG,TommerupN.ISCN2005–aninternationalsystem
ofhumancytogeneticnomenclature.Switzerland:S.Karger;
2005.
11.vanDongenJJ,MacintyreEA,GabertJA,DelabesseE,RossiV,
SaglioG,etal.StandardizedRT-PCRanalysisoffusiongene
transcriptsfromchromosomeaberrationsinacuteleukemia
fordetectionofminimalresidualdisease.Reportofthe
BIOMED-1ConcertedAction:investigationofminimal
residualdiseaseinacuteleukemia.Leukemia.
1999;13(12):1901–28.
12.JansenMW,CorralL,vanderVeldenVH,Panzer-GrümayerR,
infantacutelymphoblasticleukemiaisrelatedtothe
occurrenceandtypeofMLLgenerearrangement.Leukemia.
2007;21(4):633–41.
13.SchmidtHH,StrehlS,ThalerD,StrunkD,SillH,LinkeschW,
etal.RT-PCRandFISHanalysisofacutemyeloidleukemia
witht(8;16)(p11;p13)andchimericMOZandCBPtranscripts:
breakpointclusterregionandclinicalimplications.Leukemia.
2004;18(6):1115–21.
14.BornholdtJ,HansenJ,SteinicheT,DictorM,AntonsenA,
WolffH,etal.K-rasmutationsinsinonasalcancersin
relationtowooddustexposure.BMCCancer.2008;8:53–63.
15.YamamotoY,KiyoiH,NakanoY,SuzukiR,KoderaY,
MiyawakiS,etal.ActivatingmutationofD835withinthe
activationloopofFLT3inhumanhematologicmalignancies.
Blood.2001;97(8):2434–9.
16.NakaoM,YokotaS,IwaiT,KanekoH,HoriikeS,KashimaK,
etal.InternaltandemduplicationoftheFLT3genefoundin
acutemyeloidleukemia.Leukemia.1996;10(12):1911–8.
17.RubnitzJE,InabaH.Childhoodacutemyeloidleukemia.BrJ
Haematol.2012;159(3):259–76.
18.CatovskyD,MatutesE.Theclassificationofacuteleukemia.
Leukemia.1992;6Suppl.2:1–6.
19.HatanoK,NagaiT,MatsuyamaT,SakaguchiY,FujiwaraS,Oh
I,etal.Leukemiacellsdirectlyphagocytosebloodcellsin
AML-associatedhemophagocyticlymphohistiocytosis:acase
reportandreviewoftheliterature.ActaHaematol.
2015;133(1):98–100.
20.JekarlDW,KimM,LimJ,KimY,HanK,LeeAW,etal.CD56
antigenexpressionandhemophagocytosisofleukemiccells
inacutemyeloidleukemiawitht(16;21)(p11;q22).IntJ
Hematol.2010;92(2):306–13.
21.ByrdJC,EdenfieldWJ,ShieldsDJ,DawsonNA.Extramedullary
myeloidcelltumorsinacutenonlymphocyticleukemia:a
clinicalreview.JClinOncol.1995;13(7):1800–16.
22.KuwabaraH,NagaiM,YamaokaG,OhnishiH,KawakamiK.
SpecificskinmanifestationsinCD56positiveacutemyeloid
leukemia.JCutanPathol.1999;26(1):1–5.
23.RozmanM,CamosM,ColomerD,VillamorN,EsteveJ,Costa
D,etal.TypeIMOZ/CBP(MYST3/CREBBP)isthemost
commonchimerictranscriptinacutemyeloidleukemiawith
t(8;16)(p11;p13)translocation.GenesChromosomesCancer.
2004;40(2):140–5.
24.GilesRH,DauwerseJG,HigginsC,PetrijF,WesselsJW,
BeverstockGC,etal.DetectionofCBPrearrangementsin
acutemyelogenousleukemiawitht(8;16).Leukemia.
1997;11(12):2087–96.
25.PanagopoulosI,TorkildsenS,GorunovaL,TierensA,
TjønnfjordGE,HeimS.Comparisonbetween
karyotyping-FISH-reversetranscriptionPCRand
RNA-sequencing-fusiongeneidentificationprogramsinthe
detectionofKAT6A-CREBBPinacutemyeloidleukemia.PLoS
ONE.2014;9(5):e96570.
26.JankaGE.Hemophagocyticsyndromes.BloodRev.
2007;21(5):245–53.
27.HenterJI,HorneA,AricóM,EgelerRM,FilipovichAH,
ImashukuS,etal.HLH-2004:diagnosticandtherapeutic
guidelinesforhemophagocyticlymphohistiocytosis.Pediatr
BloodCancer.2007;48(2):124–31.
28.WongKF,YuenHL,SiuLL,PangA,KwongYL.t(8;16)(p11;p13)
predisposestoatransientbutpotentiallyrecurringneonatal
leukemia.HumPathol.2008;39(11):1702–7.
29.TeruiK,SatoT,SasakiS,KudoK,KamioT,ItoE.Twonovel
variantsofMOZ-CBPfusiontranscriptsinspontaneously
remittedinfantleukemiawitht(1;16;8)(p13;p13;p11),anew
variantoft(8;16)(p11;p13).Haematologica.2008;93(10):1591–3.
30.SainatiL,BolcatoS,CocitoMG,ZanescoL,BassoG,Montaldi
A,etal.Transientacutemonoblasticleukemiawith
reciprocal(8;16)(p11;p13)translocation.PediatrHematol
Oncol.1996;13(2):151–7.
31.CamósM,EsteveJ,JaresP,ColomerD,RozmanM,VillamorN,
etal.Geneexpressionprofilingofacutemyeloidleukemia
withtranslocationt(8;16)(p11;p13)andMYST3-CREBBP
rearrangementrevealsadistinctivesignaturewithaspecific
patternofHOXgeneexpression.CancerRes.
2006;66(14):6947–54.
32.SerravalleS,MelchiondaF,AstolfiA,LibriV,MasettiR,
PessionA.AnovelspecificsignatureofpediatricMOZ-CBP