rev bras hematol hemoter. 2017;39(4):368–371
w w w . r b h h . o r g
Revista
Brasileira
de
Hematologia
e
Hemoterapia
Brazilian
Journal
of
Hematology
and
Hemotherapy
Case
Report
DNA
microarray
expression
profiling
of
a
new
t(8;13)
AML
case
allows
identification
of
possible
leukemogenic
transformation
markers
Aline
Rangel
Pozzo
a,
Fernanda
Costas
Casal
de
Faria
a,
Luize
Otero
de
Carvalho
a,
Marcos
Barcelos
de
Pinho
b,
Raquel
Ciuvalschi
Maia
a,∗aInstitutoNacionaldeCâncer(INCA),RiodeJaneiro,RJ,Brazil
bUniversidadeFederaldoRiodeJaneiro(UFRJ),RJ,Brazil
a
r
t
i
c
l
e
i
n
f
o
Articlehistory: Received10April2017 Accepted23June2017 Availableonline22July2017
Introduction
Acutemyeloidleukemia(AML)isadiseasecharacterizedby clonalproliferationandaccumulationofmyeloidprogenitor cellsinthebonemarrow(BM),inhibitionofcell differentia-tion,increasedproliferativeindexanddefectiveapoptosis.1 AMLsecondarytomyelodysplasticsyndrome(MDS–sAML)is characterizedbydiversecytogeneticandmolecularchanges including del(5q), as well as changes in the RNA splicing pathway, TET2, EZH2, FLT3, NRAS, NPM1, RUNX1, DNMT3a, IDH1, IDH2, TET2, TP53 genes, etc.2 Established prognostic factors inAMLinclude age, the cytogeneticand molecular profileandhistory ofhematologicdisorders,suchasMDS.1 About40%ofelderlyAMLpatientswerepreviouslydiagnosed with MDS, which is usually refractory to chemotherapy.1 Wepresent a caseof asAML, which showed a singleand
∗ Correspondingauthorat:LaboratóriodeHemato-OncologiaCelulareMolecular,InstitutoNacionaldeCâncer,Prac¸adaCruzVermelha
23,6◦andar,Centro,20230-130RiodeJaneiro,RJ,Brazil. E-mailaddress:rcmaia@inca.gov.br(R.C.Maia).
previouslynotdescribedabnormality,achromosomal translo-cationbetweenchromosomes8and 13t(8;13) identifiedby conventionalcytogenetics,andseveralalteredgenesdetected by DNA microarray assay, suggesting that the t(8;13) rear-rangedregionresultsinalteredgeneexpressionpatterns.
Case
report
Thepatientwasa72-year-oldfemaleadmittedtothe Insti-tuto Nacional de Cancer(INCA), Rio deJaneiro, Brazil.She presentedwithahistoryofincreasingfatigueanddyspnea. Pastmedicalhistoryincludedarterialhypertension,coronary arterydisease,myalgiaandpneumonia.Thelaboratoryresults atdiagnosisrevealed:whitebloodcellcount36.9×109/Lwith
41%monocytoidblasts,hematocrit23%,hemoglobin7.6g/dL, platelets131.0×109/Landlactatedehydrogenase806IU/L.A
http://dx.doi.org/10.1016/j.bjhh.2017.06.003
rev bras hematol hemoter. 2017;39(4):368–371
369
bone marrow aspirate showed 34% myeloid and monocy-toidblasts(FABM4)withdysplasticfeaturesinerythroidand myeloid cells. Flowcytometry immunophenotyping identi-fiedCD33+,CD14+,CD11b+,CD13+,CD38−,HLA-DR−,CD15+,
MPO−, CD117+, TDT−, cCD79a−, CD2−, and CD7− cells.
Reversetranscription-polymerasechainreaction(RT-PCR)for RUNX1/RUNX1T1, BCR/ABL1 and MYH11/CBFB, showed no fusiontranscripts.G-bandingrevealedanabnormalkaryotype andthegeneexpressionprofilewasinvestigatedforaltered genesintherearrangedt(8;13)region.
This study was approved by the institutional review board of INCA (number 110/06). The study was conducted in accordance with the Helsinki Declaration as revised in 2008.
Methods
Cytogenetic analysis was performed on unstimulated BM cellsfor24haccordingtostandardprotocolswiththe karyo-typebeingdescribedaccording tothe InternationalSystem for Human Cytogenetic Nomenclature (ISCN, 2013).3 The geneexpressionprofilewasinvestigatedusinganAffymetrix GeneChipHumanGene1.0STArray(AffymetrixInc.,Santa Clara, CA, USA) following the manufacturer’s instructions toidentifydifferentiallyexpressedgenesin therearranged t(8;13)region.Arraydatawereextractedandprocessedwith the open softwarepackagesfrom the BioconductorProject
(www.bioconductor.org). In brief, the data was normalized
withRobustMulti-ArrayAverageexpressionmeasure. Subse-quently,anon-specificfilterwasappliedwiththegenefilter package4 in order toremove Affymetrix probes and genes that exhibited low variance across samples; differentially expressedgenes were selectedusing the linearmodels for microarray data package and summarized at log2>2. RNA extractedfromsAMLperipheralbloodwascomparedto nor-mal peripheral blood using a pool of six healthy donors. Patient’srelativegeneexpressionlevelswerecomparedtothe controlsamplelevel and listsof‘up-regulated’ and ‘down-regulated’genesweregeneratedbytheprogram.
Results
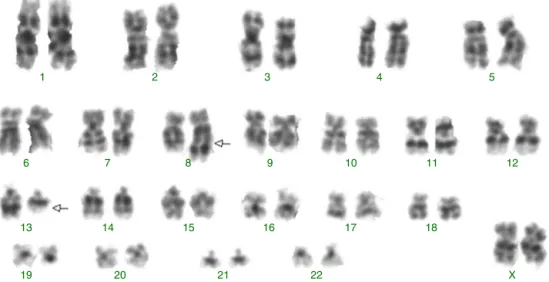
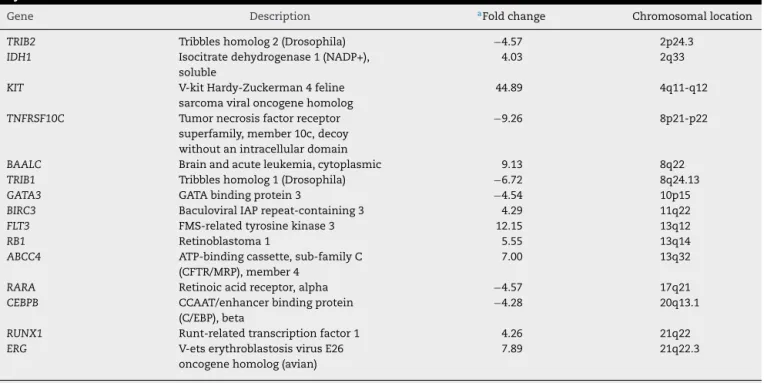
G-banding revealed an abnormal karyotype with a translocation involving chromosome 8 and 13, resulting in the previously undescribed karyotype 46,XY,t(8;13)(q22;q11)[4]/46,XX[28] (Figure 1) according to thecatalogsofcancercytogenetics.5Themicroarrayanalysis labeled874asdifferentiallyexpressedinover28,000genes. Thegeneexpressionlevels(foldchange), showninTable1, demonstrated down-regulated genes involved in myeloid development or apoptosis, including CEBPB, RARA, GATA3, TRIB1, TRIB2 and TNFRSF10C (TRAIL-R3). In addition, up-regulated genesrelated toproliferation,differentiation and drugresistance,suchasKIT,IDH1,ERG,BIRC3andABCC4were observed by microarray analysis. The gene RUNX1 (AML1) requiredforthegenerationofdefinitivehematopoieticstem cellsduringembryogenesiswasup-regulated.Thebrainand acuteleukemiacytoplasmic (BAALC)gene,locatedinthe8q22 region,andtheFLT3andRB1genes,locatedinthe13q12and 13q14 regions, respectively, appear to beintimately linked to the t(8;13). These genes were up-regulated suggesting that they could be altered due to the rearrangement of this region. Another gene that may have been inactivated duetothisrearrangementistheTRIB1gene,locatedinthe 8q24.13region,whichisapotentnegativeregulatorofMAPK signaling.
Discussion
sAMLdevelopsinapproximately 40%ofpatientswithMDS andtheclinicaldiscriminationbetweenAMLandMDSisbased oncytomorphologicalanalysis,sincepatientswithMDShave dysplastichematopoiesisandamyeloblastcountoflessthan 20%,whereasthosewithamyeloblastcountof20%ormore have AML.6 sAMLhas clinical and biological heterogeneity linkedtochromosomeaberrationsormolecularchangeswith theassociationbetweenthemsuggestingthatthose mecha-nismsaresignificantlyinvolvedinleukemogenesis.1Thiscase
1
6
13
19 20 21 22 X
14 15 16 17 18
8
7 9 10 11 12
3
2 4 5
370
rev bras hematol hemoter. 2017;39(4):368–371Table1–Up-regulatedanddown-regulatedgenesint(8;13)acutemyeloidleukemiasecondarytomyelodysplastic syndrome.
Gene Description aFoldchange Chromosomallocation
TRIB2 Tribbleshomolog2(Drosophila) −4.57 2p24.3
IDH1 Isocitratedehydrogenase1(NADP+),
soluble
4.03 2q33
KIT V-kitHardy-Zuckerman4feline
sarcomaviraloncogenehomolog
44.89 4q11-q12
TNFRSF10C Tumornecrosisfactorreceptor
superfamily,member10c,decoy withoutanintracellulardomain
−9.26 8p21-p22
BAALC Brainandacuteleukemia,cytoplasmic 9.13 8q22
TRIB1 Tribbleshomolog1(Drosophila) −6.72 8q24.13
GATA3 GATAbindingprotein3 −4.54 10p15
BIRC3 BaculoviralIAPrepeat-containing3 4.29 11q22
FLT3 FMS-relatedtyrosinekinase3 12.15 13q12
RB1 Retinoblastoma1 5.55 13q14
ABCC4 ATP-bindingcassette,sub-familyC
(CFTR/MRP),member4
7.00 13q32
RARA Retinoicacidreceptor,alpha −4.57 17q21
CEBPB CCAAT/enhancerbindingprotein
(C/EBP),beta
−4.28 20q13.1
RUNX1 Runt-relatedtranscriptionfactor1 4.26 21q22
ERG V-etserythroblastosisvirusE26
oncogenehomolog(avian)
7.89 21q22.3
a Ratiosbetweenpatientrelativegeneexpressionlevelsandcontrols.
reportshowsevidencethatt(8;13)(q22;q11)couldbeinvolved inthepathogenesisandseverityofAML.Thetranslocation t(8;13)withbreakpointsat(8q22)and(13q11)hasneitherbeen reportednordescribedforpossiblealteredgenes.Thegene expressionprofilewas performedtodeterminethe specific signatureincellsfromthispatientandtotrytoclarifyanew possiblemolecularpathwayinvolvedindiseaseevolution.Of the874genesdifferentiallyexpressed,wefocusedmainlyon genesrelatedtoAMLpathogenesis,prognosisandresponseto standardtherapy,andthegeneslocatedinthealtered chro-mosomeregion.ImportantgenessuchasCEBPB,whichplaysa pivotalroleinproliferationanddifferentiation,including sup-pressionofmyeloidleukemogenesis,7RARA,whichisrequired foroptimalmyelomonocyticdifferentiation7andGATA3 asso-ciatedwitherythropoiesis8weredown-regulatedaswasthe tumorsuppressorgeneTRIB2andTNFRSF10C(TRAIL-R3)which isimportantininitiatingapoptosis.9,10 Genesinvolvedwith anti-apoptoticfunctionsandresistancetodrugtherapysuch asBIRC3andABCC4,11,12aswellasgenesthatparticipatein theinhibitionofdifferentiationorinductionofproliferation suchas,KIT, IDH1and ERGwereup-regulated.13–15 Another genethat wasup-regulated istheRUNX1 gene,which acts asaregulatoroftheexpressionofvariousgenesspecificto hematopoiesis,andplaysanimportantroleinmyeloid differ-entiation.RUNX1amplificationisassociatedwithincreased risk of relapse and worse overall outcome in AML.16 The BAALCgene,locatedinthe 8q22.3region,isanovel molec-ularmarkerindicatinganunfavorableoutcomeinAMLwith normal cytogenetics.17 Its high expression may act as an adverse prognostic factor through prompting cell prolifer-ation and inhibiting apoptosis in leukemia cells such as in AML, acute lymphoblastic leukemia (ALL), and chronic myelogenousleukemiainblastcrisispatients.17Highlevels
oftheFLT3-wild-typereceptormaypromoteconstitutive acti-vation ofthis receptorinmalignantcells and isassociated withaworseprognosisandhighriskofrelapseinpediatric AMLpatients.18TheRB1 gene,locatedinthe13q14 region, isknownasatumorsuppressor;abnormalitiesaffectingthe RB1pathwayarenotalwaysrelatedtogeneexpression.These abnormalities areusuallyassociatedwithalackofthepRB proteinortheexpressionofamutantpRB,andinappropriate phosphorylationofpRB,leadingtoderegulatedG1-S transi-tion,thatfrequentlyoccursinmalignant disorders.19 There arealternativemechanismsthatdown-regulatepRBprotein levelsthatincludereducedtranslationoftheRB1mRNA,or reducedhalf-lifeofthemRNAandproteinshavingarolein leukemogenesis.19TheTribblesgene(TRIB-1)isapotent neg-ative regulatorofMAPKpathwaysthat influenceapoptosis, differentiation and cell-cycle progression.It isknown asa tumorsuppressorandisusuallydown-regulatedinAML.9The expressionoftheBAALC,FLT3andRB1genesinthisspecific regionappearsrelevantforthepathogenesisofAMLasthey couldberegulatedtogetherinoneregulatorymodule.Such combined expressionsuggests aprobablehigher functional relevancewhencomparedtotheexpressionofeachgene inde-pendently.
Conclusions
rev bras hematol hemoter. 2017;39(4):368–371
371
thiscaseoft(8;13).Abetterunderstandingofthemolecular processesaffectedbythefusionofthesegenesinvolvedin sAMLmayshedlightontheroleofthistranslocationin leuke-mogenesis,diseaseprogressionanditsprognosticeffects.
Contributions
ARPandRCMwereinvolvedindesigningthestudyandthe lit-eraturesearchonthesubject;RCMwasinvolvedingathering theclinicaldata;LOCwasinvolvedinperformingthe cyto-geneticanalysis;FCCFandMBPwereinvolvedinperforming geneexpressionanalysis;ARP,FCCF,LOC;MBPandRCMwere allinvolvedinwritingandeditingthemanuscript.
Conflicts
of
interest
Theauthorsdeclarenoconflictsofinterest.
Acknowledgements
Theauthors thank toMarcos Antonio M. Scheiner for his invaluable comments. This study was supported by INCT, CNPqandFAPERJ.
r
e
f
e
r
e
n
c
e
s
1. LowenbergB,DowningJR,BurnettA.Acutemyeloid
leukemia.NEnglJMed.1999;341:1051–62.
2. MilosevicJD1,PudaA,MalcovatiL,BergT,HofbauerM,
StukalovA,etal.Clinicalsignificanceofgeneticaberrations
insecondaryacutemyeloidleukemia.AmJHematol.
2012;87(11):1010–6.
3. ShafferLG,McGowan-JordanJ,SchmidM.ISCN2013:an
internationalsystemforhumancytogeneticnomenclature.
1sted.S.Karger;2013,140p.
4. SmythGK,MichaudJ,ScottHS.Useofwithin-arrayreplicate
spotsforassessingdifferentialexpressioninmicroarray
experiments.Bioinformatics.2005;21(9):2067–75.
5. MitlemanandAtlasofGeneticsandCytogeneticsinOncology andHematology,http://atlasgeneticsoncology.org/.
6. VardimanJW,ThieleJ,ArberDA,BrunningRD,BorowitzMJ,
PorwitA,etal.The2008revisionoftheWorldHealth
Organization(WHO)classificationofmyeloidneoplasmsand
acuteleukemia:rationaleandimportantchanges.Blood.
2009;114(5):937–51.
7.TruongBT,LeeYJ,LodieTA,ParkDJ,PerrottiD,WatanabeN,
etal.CCAAT/enhancerbindingproteinsrepresstheleukemic
phenotypeofacutemyeloidleukemia.Blood.
2003;101(3):1141–8.
8.VisvaderJ,AdamsJM.Megakaryocyticdifferentiationinduced
in416BmyeloidcellsbyGATA-2andGATA-3transgenesor
5-azacytidineistightlycoupledtoGATA-1expression.Blood.
1993;82(5):1493–501.
9.GilbyDC,SungHY,WinshipPR,GoodeveAC,ReillyJT,
Kiss-TothE.Tribbles-1and-2aretumoursuppressors,
down-regulatedinhumanacutemyeloidleukaemia.
ImmunolLett.2010;130(1–2):115–24.
10.RiccioniR,PasquiniL,MarianiG,SaulleE,RossiniA,Diverio
D,etal.TRAILdecoyreceptorsmediateresistanceofacute
myeloidleukemiacellstoTRAIL.Haematologica.
2005;90(5):612–24.
11.HessCJ,BerkhofJ,DenkersF,OssenkoppeleGJ,SchoutenJP,
OudejansJJ,etal.Activatedintrinsicapoptosispathwayisa
keyrelatedprognosticparameterinacutemyeloidleukemia.
JClinOncol.2007;25(10):1209–15.
12.JedlitschkyG,BurchellB,KepplerD.Themultidrugresistance
protein5functionsasanATP-dependentexportpumpfor
cyclicnucleotides.JBiolChem.2000;275:30069–74.
13.YardenY,KuangWJ,Yang-FengT,CoussensL,MunemitsuS,
DullTJ,etal.Humanproto-oncogenec-kit:anewcellsurface
receptortyrosinekinaseforanunidentifiedligand.EMBOJ.
1987;6(11):3341–51.
14.SmolkováK,Je ˇzekP.Theroleofmitochondrial
NADPH-dependentisocitratedehydrogenaseincancercells.
IntJCellBiol.2012;2012:273947.
15.MarcucciG,BaldusCD,RuppertAS,RadmacherMD,Mrózek
K,WhitmanSP,etal.OverexpressionoftheETS-relatedgene,
ERG,predictsaworseoutcomeinacutemyeloidleukemia
withnormalkaryotype:aCancerandLeukemiaGroupB
study.JClinOncol.2005;23(36):9234–42.
16.ReichardKK,KangH,RobinettS.PediatricB-lymphoblastic
leukemiawithRUNX1amplification:clinicopathologicstudy
ofeightcases.ModPathol.2011;24(12):1606–11.
17.XuB,ChenG,ShiP,GuoX,XiaoP,WangW,etal.
shRNA-mediatedBAALCknockdownaffectsproliferationand
apoptosisinhumanacutemyeloidleukemiacells.
Hematology.2012;17(1):35–40.
18.KangHJ,LeeJW,KhoSH,KimMJ,SeoYJ,KimH,etal.High
transcriptlevelofFLT3associatedwithhighriskofrelapsein
pediatricacutemyeloidleukemia.JKoreanMedSci.
2010;25(6):841–5.
19.ZhuYM,BradburyD,RussellN.Decreasedretinoblastoma
proteinexpressioninacutemyeloblasticleukaemiais
associatedwiththeautonomousproliferationofclonogenic